Glioma glycolipid metabolism: MSI2-SNORD12B-FIP1L1-ZBTB4 feedback loop as a potential treatment target
- PMID: 34047477
- PMCID: PMC8114150
- DOI: 10.1002/ctm2.411
Glioma glycolipid metabolism: MSI2-SNORD12B-FIP1L1-ZBTB4 feedback loop as a potential treatment target
Abstract
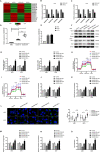
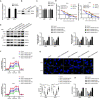
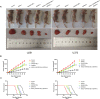
Abnormal energy metabolism, including enhanced aerobic glycolysis and lipid synthesis, is a well-established feature of glioblastoma (GBM) cells. Thus, targeting the cellular glycolipid metabolism can be a feasible therapeutic strategy for GBM. This study aimed to evaluate the roles of MSI2, SNORD12B, and ZBTB4 in regulating the glycolipid metabolism and proliferation of GBM cells. MSI2 and SNORD12B expression was significantly upregulated and ZBTB4 expression was significantly low in GBM tissues and cells. Knockdown of MSI2 or SNORD12B or overexpression of ZBTB4 inhibited GBM cell glycolipid metabolism and proliferation. MSI2 may improve SNORD12B expression by increasing its stability. Importantly, SNORD12B increased utilization of the ZBTB4 mRNA transcript distal polyadenylation signal in alternative polyadenylation processing by competitively combining with FIP1L1, which decreased ZBTB4 expression because of the increased proportion of the 3' untranslated region long transcript. ZBTB4 transcriptionally suppressed the expression of HK2 and ACLY by binding directly to the promoter regions. Additionally, ZBTB4 bound the MSI promoter region to transcriptionally suppress MSI2 expression, thereby forming an MSI2/SNORD12B/FIP1L1/ZBTB4 feedback loop to regulate the glycolipid metabolism and proliferation of GBM cells. In conclusion, MSI2 increased the stability of SNORD12B, which regulated ZBTB4 alternative polyadenylation processing by competitively binding to FIP1L1. Thus, the MSI2/SNORD12B/FIP1L1/ZBTB4 positive feedback loop plays a crucial role in regulating the glycolipid metabolism of GBM cells and provides a potential drug target for glioma treatment.
Keywords: MSI2; SNORD12B; ZBTB4; alternative polyadenylation; glioma; glycolipid metabolism; molecular pattern; potential target; prediction and personalized prevention; translational research.
© 2021 The Authors. Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare that there is no conflict of interest.
Figures





Similar articles
-
Effect of SNORD113-3/ADAR2 on glycolipid metabolism in glioblastoma via A-to-I editing of PHKA2.Cell Mol Biol Lett. 2025 Jan 10;30(1):5. doi: 10.1186/s11658-024-00680-9. Cell Mol Biol Lett. 2025. PMID: 39794701 Free PMC article.
-
U3 snoRNA-mediated degradation of ZBTB7A regulates aerobic glycolysis in isocitrate dehydrogenase 1 wild-type glioblastoma cells.CNS Neurosci Ther. 2023 Oct;29(10):2811-2825. doi: 10.1111/cns.14218. Epub 2023 Apr 17. CNS Neurosci Ther. 2023. PMID: 37066523 Free PMC article.
-
MSI2-TGF-β/TGF-β R1/SMAD3 positive feedback regulation in glioblastoma.Cancer Chemother Pharmacol. 2019 Aug;84(2):415-425. doi: 10.1007/s00280-019-03892-5. Epub 2019 Jun 27. Cancer Chemother Pharmacol. 2019. PMID: 31250154
-
Harnessing OLIG2 function in tumorigenicity and plasticity to target malignant gliomas.Cell Cycle. 2017 Sep 17;16(18):1654-1660. doi: 10.1080/15384101.2017.1361062. Epub 2017 Aug 14. Cell Cycle. 2017. PMID: 28806136 Free PMC article. Review.
-
Modulation of untranslated region alternative polyadenylation in glioma tumorigenesis.Biomed Pharmacother. 2021 May;137:111416. doi: 10.1016/j.biopha.2021.111416. Epub 2021 Feb 24. Biomed Pharmacother. 2021. PMID: 33761620 Review.
Cited by
-
Characterization of circRNA-Associated-ceRNA Networks Involved in the Pathogenesis of Postoperative Cognitive Dysfunction in Aging Mice.Front Aging Neurosci. 2022 Apr 4;14:727805. doi: 10.3389/fnagi.2022.727805. eCollection 2022. Front Aging Neurosci. 2022. PMID: 35444525 Free PMC article.
-
Effect of SNORD113-3/ADAR2 on glycolipid metabolism in glioblastoma via A-to-I editing of PHKA2.Cell Mol Biol Lett. 2025 Jan 10;30(1):5. doi: 10.1186/s11658-024-00680-9. Cell Mol Biol Lett. 2025. PMID: 39794701 Free PMC article.
-
Detection of a Novel MSI2-C17orf64 Transcript in a Patient with Aggressive Adenocarcinoma of the Gastroesophageal Junction: A Case Report.Genes (Basel). 2023 Apr 15;14(4):918. doi: 10.3390/genes14040918. Genes (Basel). 2023. PMID: 37107676 Free PMC article.
-
Musashi-2 potentiates colorectal cancer immune infiltration by regulating the post-translational modifications of HMGB1 to promote DCs maturation and migration.Cell Commun Signal. 2024 Feb 12;22(1):117. doi: 10.1186/s12964-024-01495-z. Cell Commun Signal. 2024. PMID: 38347600 Free PMC article.
-
SNORD17-mediated KAT6B mRNA 2'-O-methylation regulates vasculogenic mimicry in glioblastoma cells.Cell Biol Toxicol. 2023 Dec;39(6):2841-2860. doi: 10.1007/s10565-023-09805-w. Epub 2023 Apr 14. Cell Biol Toxicol. 2023. PMID: 37058271
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
