ESCRT, not intralumenal fragments, sorts ubiquitinated vacuole membrane proteins for degradation
- PMID: 34047770
- PMCID: PMC8167898
- DOI: 10.1083/jcb.202012104
ESCRT, not intralumenal fragments, sorts ubiquitinated vacuole membrane proteins for degradation
Abstract
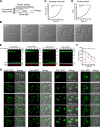
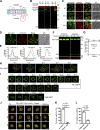
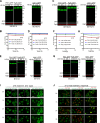
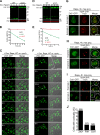
The lysosome (or vacuole in fungi and plants) is an essential organelle for nutrient sensing and cellular homeostasis. In response to environmental stresses such as starvation, the yeast vacuole can adjust its membrane composition by selectively internalizing membrane proteins into the lumen for degradation. Regarding the selective internalization mechanism, two competing models have been proposed. One model suggests that the ESCRT machinery is responsible for the sorting. In contrast, the ESCRT-independent intralumenal fragment (ILF) pathway proposes that the fragment generated by homotypic vacuole fusion is responsible for the sorting. Here, we applied a microfluidics-based imaging method to capture the complete degradation process in vivo. Combining live-cell imaging with a synchronized ubiquitination system, we demonstrated that ILF cargoes are not degraded through intralumenal fragments. Instead, ESCRTs function on the vacuole membrane to sort them into the lumen for degradation. We further discussed challenges in reconstituting vacuole membrane protein degradation.
© 2021 Yang et al.
Figures




Comment in
-
How the vacuole ESCRTs its own proteins to their final destination.J Cell Biol. 2021 Aug 2;220(8):e202105177. doi: 10.1083/jcb.202105177. Epub 2021 Jul 19. J Cell Biol. 2021. PMID: 34279549 Free PMC article.
References
-
- Adell, M.A.Y., Migliano S.M., Upadhyayula S., Bykov Y.S., Sprenger S., Pakdel M., Vogel G.F., Jih G., Skillern W., Behrouzi R., et al. 2017. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife. 6:e31652. 10.7554/eLife.31652 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

