Effects of cofD gene knock-out on the methanogenesis of Methanobrevibacter ruminantium
- PMID: 34047886
- PMCID: PMC8163928
- DOI: 10.1186/s13568-021-01236-2
Effects of cofD gene knock-out on the methanogenesis of Methanobrevibacter ruminantium
Abstract
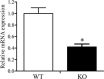
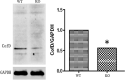
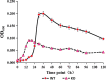
This study aimed to investigate the effects of cofD gene knock-out on the synthesis of coenzyme F420 and production of methane in Methanobrevibacter ruminantium (M. ruminantium). The experiment successfully constructed a cofD gene knock-out M. ruminantium via homologous recombination technology. The results showed that the logarithmic phase of mutant M. ruminantium (12 h) was lower than the wild-type (24 h). The maximum biomass and specific growth rate of mutant M. ruminantium were significantly lower (P < 0.05) than those of wild-type, and the maximum biomass of mutant M. ruminantium was approximately half of the wild-type; meanwhile, the proliferation was reduced. The synthesis amount of coenzyme F420 of M. ruminantium was significantly decreased (P < 0.05) after the cofD gene knock-out. Moreover, the maximum amount of H2 consumed and CH4 produced by mutant were 14 and 2% of wild-type M. ruminantium respectively. In conclusion, cofD gene knock-out induced the decreased growth rate and reproductive ability of M. ruminantium. Subsequently, the synthesis of coenzyme F420 was decreased. Ultimately, the production capacity of CH4 in M. ruminantium was reduced. Our research provides evidence that cofD gene plays an indispensable role in the regulation of coenzyme F420 synthesis and CH4 production in M. ruminantium.
Keywords: Coenzyme F420; Gene knock-out; M ethanobrevibacter ruminantium; Methane; cofD gene.
Conflict of interest statement
The authors declare no competing interests.
Figures
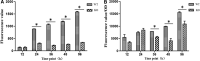
Similar articles
-
Diversification by CofC and Control by CofD Govern Biosynthesis and Evolution of Coenzyme F420 and Its Derivative 3PG-F420.mBio. 2022 Feb 22;13(1):e0350121. doi: 10.1128/mbio.03501-21. Epub 2022 Jan 18. mBio. 2022. PMID: 35038903 Free PMC article.
-
Hydrogen regulation of growth, growth yields, and methane gene transcription in Methanobacterium thermoautotrophicum deltaH.J Bacteriol. 1997 Feb;179(3):889-98. doi: 10.1128/jb.179.3.889-898.1997. J Bacteriol. 1997. PMID: 9006047 Free PMC article.
-
Growth Kinetics, Carbon Isotope Fractionation, and Gene Expression in the Hyperthermophile Methanocaldococcus jannaschii during Hydrogen-Limited Growth and Interspecies Hydrogen Transfer.Appl Environ Microbiol. 2019 Apr 18;85(9):e00180-19. doi: 10.1128/AEM.00180-19. Print 2019 May 1. Appl Environ Microbiol. 2019. PMID: 30824444 Free PMC article.
-
Electron Bifurcation and Confurcation in Methanogenesis and Reverse Methanogenesis.Front Microbiol. 2018 Jun 20;9:1322. doi: 10.3389/fmicb.2018.01322. eCollection 2018. Front Microbiol. 2018. PMID: 29973922 Free PMC article. Review.
-
The unique biochemistry of methanogenesis.Prog Nucleic Acid Res Mol Biol. 2002;71:223-83. doi: 10.1016/s0079-6603(02)71045-3. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102556 Review.
Cited by
-
Teaching old dogs new tricks: genetic engineering methanogens.Appl Environ Microbiol. 2024 Jul 24;90(7):e0224723. doi: 10.1128/aem.02247-23. Epub 2024 Jun 10. Appl Environ Microbiol. 2024. PMID: 38856201 Free PMC article. Review.
References
-
- André-Denis GW, Klieve AV. Does the complexity of the rumen microbial ecology preclude methane mitigation? Anim Feed Sci Technol. 2011;166:248–253.
-
- Benepal PK (2012) The rumen methanogen community and diurnal activity in pasture based dairy cows of the South Island. Doctoral thesis, Lincoln University, New Zealand
-
- Choi KP, Bair TB, Bae YM, Daniels L. Use of Transposon Tn5367 Mutagenesis and a Nitroimidazopyran-Based selection system to demonstrate a requirement for fbiA and fbiB in Coenzyme F420 Biosynthesis by Mycobacterium bovis BCG. J Bacteriol. 2001;183(24):7058–7066. doi: 10.1128/JB.183.24.7058-7066.2001. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

