The DNMT3A PWWP domain is essential for the normal DNA methylation landscape in mouse somatic cells and oocytes
- PMID: 34048432
- PMCID: PMC8162659
- DOI: 10.1371/journal.pgen.1009570
The DNMT3A PWWP domain is essential for the normal DNA methylation landscape in mouse somatic cells and oocytes
Abstract
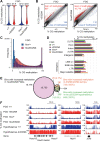
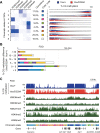
DNA methylation at CG sites is important for gene regulation and embryonic development. In mouse oocytes, de novo CG methylation requires preceding transcription-coupled histone mark H3K36me3 and is mediated by a DNA methyltransferase DNMT3A. DNMT3A has a PWWP domain, which recognizes H3K36me2/3, and heterozygous mutations in this domain, including D329A substitution, cause aberrant CG hypermethylation of regions marked by H3K27me3 in somatic cells, leading to a dwarfism phenotype. We herein demonstrate that D329A homozygous mice show greater CG hypermethylation and severer dwarfism. In oocytes, D329A substitution did not affect CG methylation of H3K36me2/3-marked regions, including maternally methylated imprinting control regions; rather, it caused aberrant hypermethylation in regions lacking H3K36me2/3, including H3K27me3-marked regions. Thus, the role of the PWWP domain in CG methylation seems similar in somatic cells and oocytes; however, there were cell-type-specific differences in affected regions. The major satellite repeat was also hypermethylated in mutant oocytes. Contrary to the CA hypomethylation in somatic cells, the mutation caused hypermethylation at CH sites, including CA sites. Surprisingly, oocytes expressing only the mutated protein could support embryonic and postnatal development. Our study reveals that the DNMT3A PWWP domain is important for suppressing aberrant CG hypermethylation in both somatic cells and oocytes but that D329A mutation has little impact on the developmental potential of oocytes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

