Duodenases are a small subfamily of ruminant intestinal serine proteases that have undergone a remarkable diversification in cleavage specificity
- PMID: 34048501
- PMCID: PMC8162674
- DOI: 10.1371/journal.pone.0252624
Duodenases are a small subfamily of ruminant intestinal serine proteases that have undergone a remarkable diversification in cleavage specificity
Abstract
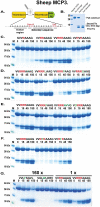
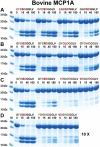
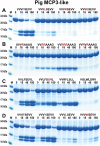
Ruminants have a very complex digestive system adapted for the digestion of cellulose rich food. Gene duplications have been central in the process of adapting their digestive system for this complex food source. One of the new loci involved in food digestion is the lysozyme c locus where cows have ten active such genes compared to a single gene in humans and where four of the bovine copies are expressed in the abomasum, the real stomach. The second locus that has become part of the ruminant digestive system is the chymase locus. The chymase locus encodes several of the major hematopoietic granule proteases. In ruminants, genes within the chymase locus have duplicated and some of them are expressed in the duodenum and are therefore called duodenases. To obtain information on their specificities and functions we produced six recombinant proteolytically active duodenases (three from cows, two from sheep and one from pigs). Two of the sheep duodenases were found to be highly specific tryptases and one of the bovine duodenases was a highly specific asp-ase. The remaining two bovine duodenases were dual enzymes with potent tryptase and chymase activities. In contrast, the pig enzyme was a chymase with no tryptase or asp-ase activity. These results point to a remarkable flexibility in both the primary and extended specificities within a single chromosomal locus that most likely has originated from one or a few genes by several rounds of local gene duplications. Interestingly, using the consensus cleavage site for the bovine asp-ase to screen the entire bovine proteome, it revealed Mucin-5B as one of the potential targets. Using the same strategy for one of the sheep tryptases, this enzyme was found to have potential cleavage sites in two chemokine receptors, CCR3 and 7, suggesting a role for this enzyme to suppress intestinal inflammation.
Conflict of interest statement
The authors have read the journal’s policy and the authors of this manuscript have the following competing interests: LG is a paid employee of GDL Pharmaceutical Consulting and Contracting and JK is a paid employee of Tosoh Bioscience LLC. There are no patents, products in development or marketing products to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials.
Figures









Similar articles
-
The Evolutionary History of the Chymase Locus -a Locus Encoding Several of the Major Hematopoietic Serine Proteases.Int J Mol Sci. 2021 Oct 11;22(20):10975. doi: 10.3390/ijms222010975. Int J Mol Sci. 2021. PMID: 34681635 Free PMC article. Review.
-
Expansion of the mast cell chymase locus over the past 200 million years of mammalian evolution.Immunogenetics. 2006 Aug;58(8):655-69. doi: 10.1007/s00251-006-0126-1. Epub 2006 Jun 29. Immunogenetics. 2006. PMID: 16807745
-
Two granzyme A/K homologs in Zebra mbuna have different specificities, one classical tryptase and one with chymase activity.Dev Comp Immunol. 2023 Nov;148:104920. doi: 10.1016/j.dci.2023.104920. Epub 2023 Aug 18. Dev Comp Immunol. 2023. PMID: 37597699
-
Extended Cleavage Specificities of Rabbit and Guinea Pig Mast Cell Chymases: Two Highly Specific Leu-Ases.Int J Mol Sci. 2019 Dec 16;20(24):6340. doi: 10.3390/ijms20246340. Int J Mol Sci. 2019. PMID: 31888202 Free PMC article.
-
Granule proteases of hematopoietic cells, a family of versatile inflammatory mediators - an update on their cleavage specificity, in vivo substrates, and evolution.Biol Chem. 2014 Jan;395(1):15-49. doi: 10.1515/hsz-2013-0211. Biol Chem. 2014. PMID: 23969467 Review.
Cited by
-
Alterations in ileal transcriptomics during an intestinal barrier challenge in lactating Holstein cows fed a Saccharomyces cerevisiae fermentation product identify potential regulatory processes.J Anim Sci. 2023 Jan 3;101:skad277. doi: 10.1093/jas/skad277. J Anim Sci. 2023. PMID: 37616596 Free PMC article.
-
Extended Cleavage Specificity of two Hematopoietic Serine Proteases from a Ray-Finned Fish, the Spotted Gar (Lepisosteus oculatus).Int J Mol Sci. 2024 Jan 30;25(3):1669. doi: 10.3390/ijms25031669. Int J Mol Sci. 2024. PMID: 38338947 Free PMC article.
-
The Evolutionary History of the Chymase Locus -a Locus Encoding Several of the Major Hematopoietic Serine Proteases.Int J Mol Sci. 2021 Oct 11;22(20):10975. doi: 10.3390/ijms222010975. Int J Mol Sci. 2021. PMID: 34681635 Free PMC article. Review.
-
The Extended Cleavage Specificity of Channel Catfish Granzyme-like II, A Highly Specific Elastase, Expressed by Natural Killer-like Cells.Int J Mol Sci. 2023 Dec 26;25(1):356. doi: 10.3390/ijms25010356. Int J Mol Sci. 2023. PMID: 38203526 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

