SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2
- PMID: 34048708
- PMCID: PMC8106883
- DOI: 10.1016/j.immuni.2021.05.006
SARS-CoV-2 exacerbates proinflammatory responses in myeloid cells through C-type lectin receptors and Tweety family member 2
Abstract
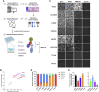
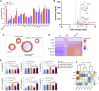
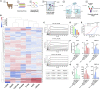
Despite mounting evidence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) engagement with immune cells, most express little, if any, of the canonical receptor of SARS-CoV-2, angiotensin-converting enzyme 2 (ACE2). Here, using a myeloid cell receptor-focused ectopic expression screen, we identified several C-type lectins (DC-SIGN, L-SIGN, LSECtin, ASGR1, and CLEC10A) and Tweety family member 2 (TTYH2) as glycan-dependent binding partners of the SARS-CoV-2 spike. Except for TTYH2, these molecules primarily interacted with spike via regions outside of the receptor-binding domain. Single-cell RNA sequencing analysis of pulmonary cells from individuals with coronavirus disease 2019 (COVID-19) indicated predominant expression of these molecules on myeloid cells. Although these receptors do not support active replication of SARS-CoV-2, their engagement with the virus induced robust proinflammatory responses in myeloid cells that correlated with COVID-19 severity. We also generated a bispecific anti-spike nanobody that not only blocked ACE2-mediated infection but also the myeloid receptor-mediated proinflammatory responses. Our findings suggest that SARS-CoV-2-myeloid receptor interactions promote immune hyperactivation, which represents potential targets for COVID-19 therapy.
Keywords: ASGR1; CLEC10A; COVID-19; DC-SIGN; L-SIGN; LSECtin; SARS-CoV-2; TTYH2; myeloid cells; proinflammatory responses.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.W., Q.L., J.L., B.H., J.D., and Y.L. are named inventors on a patent application that describes the anti-SARS-CoV-2 blocking nanobodies. J.W. is a consultant for Lilly Asia Ventures and is on the Scientific Advisory Board of Rootpath Genomics, which is not relevant to this work. M.S.D. is a consultant for Inbios, Vir Biotechnology, and NGM Biopharmaceuticals and is on the Scientific Advisory Board of Moderna and Immunome. The Diamond laboratory has received unrelated funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI059167/AI/NIAID NIH HHS/United States
- R21 AI163924/AI/NIAID NIH HHS/United States
- R00 AI135031/AI/NIAID NIH HHS/United States
- T32 AI007517/AI/NIAID NIH HHS/United States
- R01 CA228135/CA/NCI NIH HHS/United States
- R01 AI150796/AI/NIAID NIH HHS/United States
- R01 CA202025/CA/NCI NIH HHS/United States
- R01 CA242020/CA/NCI NIH HHS/United States
- P30 DK052574/DK/NIDDK NIH HHS/United States
- UL1 TR001863/TR/NCATS NIH HHS/United States
- R01 CA202027/CA/NCI NIH HHS/United States
- R01 AI157155/AI/NIAID NIH HHS/United States
- R01 AI143861/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

