Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients
- PMID: 34048985
- PMCID: PMC8149473
- DOI: 10.1016/j.trsl.2021.05.004
Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients
Abstract
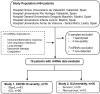
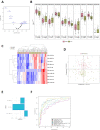
We aimed to examine the circulating microRNA (miRNA) profile of hospitalized COVID-19 patients and evaluate its potential as a source of biomarkers for the management of the disease. This was an observational and multicenter study that included 84 patients with a positive nasopharyngeal swab Polymerase chain reaction (PCR) test for SARS-CoV-2 recruited during the first pandemic wave in Spain (March-June 2020). Patients were stratified according to disease severity: hospitalized patients admitted to the clinical wards without requiring critical care and patients admitted to the intensive care unit (ICU). An additional study was completed including ICU nonsurvivors and survivors. Plasma miRNA profiling was performed using reverse transcription polymerase quantitative chain reaction (RT-qPCR). Predictive models were constructed using least absolute shrinkage and selection operator (LASSO) regression. Ten circulating miRNAs were dysregulated in ICU patients compared to ward patients. LASSO analysis identified a signature of three miRNAs (miR-148a-3p, miR-451a and miR-486-5p) that distinguishes between ICU and ward patients [AUC (95% CI) = 0.89 (0.81-0.97)]. Among critically ill patients, six miRNAs were downregulated between nonsurvivors and survivors. A signature based on two miRNAs (miR-192-5p and miR-323a-3p) differentiated ICU nonsurvivors from survivors [AUC (95% CI) = 0.80 (0.64-0.96)]. The discriminatory potential of the signature was higher than that observed for laboratory parameters such as leukocyte counts, C-reactive protein (CRP) or D-dimer [maximum AUC (95% CI) for these variables = 0.73 (0.55-0.92)]. miRNA levels were correlated with the duration of ICU stay. Specific circulating miRNA profiles are associated with the severity of COVID-19. Plasma miRNA signatures emerge as a novel tool to assist in the early prediction of vital status deterioration among ICU patients.
Copyright © 2021 Elsevier Inc. All rights reserved.
Figures

References
-
- Lin B, Liu J, Liu Y, Qin X. Progress in understanding COVID-19: insights from the omics approach. Crit Rev Clin Lab Sci. 2020;58(4):1–18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

