Comparing infectivity and virulence of emerging SARS-CoV-2 variants in Syrian hamsters
- PMID: 34049240
- PMCID: PMC8143995
- DOI: 10.1016/j.ebiom.2021.103403
Comparing infectivity and virulence of emerging SARS-CoV-2 variants in Syrian hamsters
Abstract
Background: Within one year after its emergence, more than 108 million people acquired SARS-CoV-2 and almost 2·4 million succumbed to COVID-19. New SARS-CoV-2 variants of concern (VoC) are emerging all over the world, with the threat of being more readily transmitted, being more virulent, or escaping naturally acquired and vaccine-induced immunity. At least three major prototypic VoC have been identified, i.e. the United Kingdom, UK (B.1.1.7), South African (B.1.351) and Brazilian (B.1.1.28.1) variants. These are replacing formerly dominant strains and sparking new COVID-19 epidemics.
Methods: We studied the effect of infection with prototypic VoC from both B.1.1.7 and B.1.351 variants in female Syrian golden hamsters to assess their relative infectivity and virulence in direct comparison to two basal SARS-CoV-2 strains isolated in early 2020.
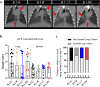
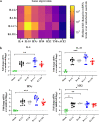
Findings: A very efficient infection of the lower respiratory tract of hamsters by these VoC is observed. In line with clinical evidence from patients infected with these VoC, no major differences in disease outcome were observed as compared to the original strains as was quantified by (i) histological scoring, (ii) micro-computed tomography, and (iii) analysis of the expression profiles of selected antiviral and pro-inflammatory cytokine genes. Noteworthy however, in hamsters infected with VoC B.1.1.7, a particularly strong elevation of proinflammatory cytokines was detected.
Interpretation: We established relevant preclinical infection models that will be pivotal to assess the efficacy of current and future vaccine(s) (candidates) as well as therapeutics (small molecules and antibodies) against two important SARS-CoV-2 VoC.
Funding: Stated in the acknowledgment.
Keywords: Emergence; Hamster model; SARS-CoV-2; Variants of concern (VoC).
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest Authors declare no conflict of interests.
Figures


References
-
- O'Toole Á., Hill V., Pybus O.G. Tracking the international spread of SARS-CoV-2 lineages B.1.1.7 and B.1.351/501Y-V2. Virological. 2021 Published online January 14. https://virological.org/t/tracking-the-international-spread-of-sars-cov-... (accessed Feb 24, 2021) - PMC - PubMed
-
- O'Toole Á., Hill V. Lineage B.1.1.7. 2021; published online February 25. https://cov-lineages.org/global_report_B.1.1.7.html (accessed Feb 26, 2021).
-
- O'Toole Á., Hill V. Lineage B.1.351. 2021; published online February 25. https://cov-lineages.org/global_report_B.1.351.html (accessed Feb 26, 2021).
-
- O'Toole Á., Hill V. Lineage P.1. 2021; published online February 25. https://cov-lineages.org/global_report_P.1.html (accessed Feb 26, 2021).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

