LncRNA:DNA triplex-forming sites are positioned at specific areas of genome organization and are predictors for Topologically Associated Domains
- PMID: 34049493
- PMCID: PMC8164242
- DOI: 10.1186/s12864-021-07727-7
LncRNA:DNA triplex-forming sites are positioned at specific areas of genome organization and are predictors for Topologically Associated Domains
Abstract
Background: Chromosomes are organized into units called topologically associated domains (TADs). TADs dictate regulatory landscapes and other DNA-dependent processes. Even though various factors that contribute to the specification of TADs have been proposed, the mechanism is not fully understood. Understanding the process for specification and maintenance of these units is essential in dissecting cellular processes and disease mechanisms.
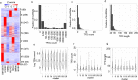
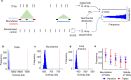
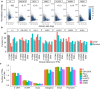
Results: In this study, we report a genome-wide study that considers the idea of long noncoding RNAs (lncRNAs) mediating chromatin organization using lncRNA:DNA triplex-forming sites (TFSs). By analyzing the TFSs of expressed lncRNAs in multiple cell lines, we find that they are enriched in TADs, their boundaries, and loop anchors. However, they are evenly distributed across different regions of a TAD showing no preference for any specific portions within TADs. No relationship is observed between the locations of these TFSs and CTCF binding sites. However, TFSs are located not just in promoter regions but also in intronic, intergenic, and 3'UTR regions. We also show these triplex-forming sites can be used as predictors in machine learning models to discriminate TADs from other genomic regions. Finally, we compile a list of important "TAD-lncRNAs" which are top predictors for TADs identification.
Conclusions: Our observations advocate the idea that lncRNA:DNA TFSs are positioned at specific areas of the genome organization and are important predictors for TADs. LncRNA:DNA triplex formation most likely is a general mechanism of action exhibited by some lncRNAs, not just for direct gene regulation but also to mediate 3D chromatin organization.
Keywords: CTCF; Long noncoding RNAs; RNA:DNA triplex; TAD-lncRNAs; TADs; Triplex structures.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Association between Triplex-Forming Sites of Cardiac Long Noncoding RNA GATA6-AS1 and Chromatin Organization.Noncoding RNA. 2022 Jun 1;8(3):41. doi: 10.3390/ncrna8030041. Noncoding RNA. 2022. PMID: 35736638 Free PMC article.
-
Genome-wide computational analysis of potential long noncoding RNA mediated DNA:DNA:RNA triplexes in the human genome.J Transl Med. 2017 Sep 2;15(1):186. doi: 10.1186/s12967-017-1282-9. J Transl Med. 2017. PMID: 28865451 Free PMC article.
-
TADKB: Family classification and a knowledge base of topologically associating domains.BMC Genomics. 2019 Mar 14;20(1):217. doi: 10.1186/s12864-019-5551-2. BMC Genomics. 2019. PMID: 30871473 Free PMC article.
-
RNA-DNA Triplex Formation by Long Noncoding RNAs.Cell Chem Biol. 2016 Nov 17;23(11):1325-1333. doi: 10.1016/j.chembiol.2016.09.011. Epub 2016 Oct 20. Cell Chem Biol. 2016. PMID: 27773629 Review.
-
TADs: Dynamic structures to create stable regulatory functions.Curr Opin Struct Biol. 2023 Aug;81:102622. doi: 10.1016/j.sbi.2023.102622. Epub 2023 Jun 9. Curr Opin Struct Biol. 2023. PMID: 37302180 Review.
Cited by
-
Dysregulation of murine long noncoding single-cell transcriptome in nonalcoholic steatohepatitis and liver fibrosis.RNA. 2023 Jul;29(7):977-1006. doi: 10.1261/rna.079580.123. Epub 2023 Apr 4. RNA. 2023. PMID: 37015806 Free PMC article.
-
Integrated lncRNA function upon genomic and epigenomic regulation.Mol Cell. 2022 Jun 16;82(12):2252-2266. doi: 10.1016/j.molcel.2022.05.027. Mol Cell. 2022. PMID: 35714586 Free PMC article. Review.
-
Quantitative estimates of the regulatory influence of long non-coding RNAs on global gene expression variation using TCGA breast cancer transcriptomic data.PLoS Comput Biol. 2024 Jun 5;20(6):e1012103. doi: 10.1371/journal.pcbi.1012103. eCollection 2024 Jun. PLoS Comput Biol. 2024. PMID: 38838009 Free PMC article.
-
Systematic study of hybrid triplex topology and stability suggests a general triplex-mediated regulatory mechanism.Nucleic Acids Res. 2025 Feb 27;53(5):gkaf170. doi: 10.1093/nar/gkaf170. Nucleic Acids Res. 2025. PMID: 40071936 Free PMC article.
-
Repetitive DNA is Functional and Encodes Parts of the Non-Coding RNA Repertoire.Adv Genet (Hoboken). 2022 Nov 26;3(4):2200026. doi: 10.1002/ggn2.202200026. eCollection 2022 Dec. Adv Genet (Hoboken). 2022. PMID: 36911293 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

