Rapid separation of blood plasma exosomes from low-density lipoproteins via a hydrophobic interaction chromatography method on a polyester capillary-channeled polymer fiber phase
- PMID: 34049630
- PMCID: PMC8164660
- DOI: 10.1016/j.aca.2021.338578
Rapid separation of blood plasma exosomes from low-density lipoproteins via a hydrophobic interaction chromatography method on a polyester capillary-channeled polymer fiber phase
Abstract
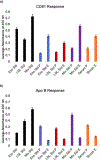
Exosomes are membrane-bound, cell-secreted vesicles, with sizes ranging from 30 to 150 nm. Exosomes in blood plasma have become proposed targets as measurable indicators of disease conditions. Current methods for plasma-based exosome isolation are time-consuming, complex, and have high operational costs. One of the most commonly reported shortcomings of current isolation protocols is the co-extraction of lipoproteins (e.g. low-density lipoproteins, LDLs) with the target exosomes. This report describes the use of a rapid, single-operation hydrophobic interaction chromatography (HIC) procedure on a polyester (PET) capillary-channeled polymer (C-CP) fiber column, demonstrating the ability to efficiently purify exosomes. The method has previously been demonstrated for isolation of exosomes from diverse biological matrices, but questions were raised about the potential co-elution of LDLs. In the method described herein, a step-gradient procedure sequentially elutes spiked lipoproteins and blood plasma-originating exosomes in 10 min, with the LDLs excluded from the desired exosome fraction. Mass spectrometry (MS) was used to characterize an impurity in the primary LDL material, identifying the presence of exosomal material. Transmission electron microscopy (TEM) and an enzyme-linked immunosorbent assay (ELISA) were used to identify the various elution components. The method serves both as a rapid means of high purity exosome isolation as well as a screening tool for the purity of LDL samples with respect to extracellular vesicles.
Keywords: Capillary-channeled polymer (C-CP); Enzyme-linked immunosorbent assay (ELISA); Exosomes; Extracellular vesicles (EVs); Fibers; Hydrophobic interaction chromatography; Low-density lipoprotein (LDL); Proteomics; Transmission electron microscopy (TEM).
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

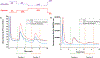

References
-
- Colombo M, Raposo G, Thery C, Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles, Annu. Rev. Cell Dev. Biol 30 (2014) 255–289. - PubMed
-
- Chaput N, Thery C, Exosomes: immune properties and potential clinical implementations, Semin Immunopathol. 33(5) (2011) 419–440. - PubMed
-
- Schneider A, Exosomes: novel biomarkers for the clinical diagnosis of neurodegenerative diseases, J. Neural Transm 126(5) (2019) 646–646.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

