Gene-Environment Interactions in Multiple Sclerosis: A UK Biobank Study
- PMID: 34049995
- PMCID: PMC8192056
- DOI: 10.1212/NXI.0000000000001007
Gene-Environment Interactions in Multiple Sclerosis: A UK Biobank Study
Abstract
Objective: We sought to determine whether genetic risk modifies the effect of environmental risk factors for multiple sclerosis (MS). To test this hypothesis, we tested for statistical interaction between polygenic risk scores (PRS) capturing genetic susceptibility to MS and environmental risk factors for MS in UK Biobank.
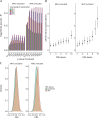
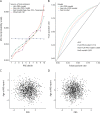
Methods: People with MS were identified within UK Biobank using ICD-10-coded MS or self-report. Associations between environmental risk factors and MS risk were quantified with a case-control design using multivariable logistic regression. PRS were derived using the clumping-and-thresholding approach with external weights from the largest genome-wide association study of MS. Separate scores were created including major histocompatibility complex (MHC) (PRSMHC) and excluding (PRSnon-MHC) the MHC locus. The best-performing PRS were identified in 30% of the cohort and validated in the remaining 70%. Interaction between environmental and genetic risk factors was quantified using the attributable proportion due to interaction (AP) and multiplicative interaction.
Results: Data were available for 2,250 people with MS and 486,000 controls. Childhood obesity, earlier age at menarche, and smoking were associated with MS. The optimal PRS were strongly associated with MS in the validation cohort (PRSMHC: Nagelkerke's pseudo-R2 0.033, p = 3.92 × 10-111; PRSnon-MHC: Nagelkerke's pseudo-R2 0.013, p = 3.73 × 10-43). There was strong evidence of interaction between polygenic risk for MS and childhood obesity (PRSMHC: AP = 0.17, 95% CI 0.06-0.25, p = 0.004; PRSnon-MHC: AP = 0.17, 95% CI 0.06-0.27, p = 0.006).
Conclusions: This study provides novel evidence for an interaction between childhood obesity and a high burden of autosomal genetic risk. These findings may have significant implications for our understanding of MS biology and inform targeted prevention strategies.
Copyright © 2021 The Author(s). Published by Wolters Kluwer Health, Inc. on behalf of the American Academy of Neurology.
Figures


Similar articles
-
No evidence for an association between alcohol consumption and Multiple Sclerosis risk: a UK Biobank study.Sci Rep. 2022 Dec 22;12(1):22158. doi: 10.1038/s41598-022-26409-2. Sci Rep. 2022. PMID: 36550182 Free PMC article.
-
Parkinson's disease determinants, prediction and gene-environment interactions in the UK Biobank.J Neurol Neurosurg Psychiatry. 2020 Oct;91(10):1046-1054. doi: 10.1136/jnnp-2020-323646. J Neurol Neurosurg Psychiatry. 2020. PMID: 32934108 Free PMC article.
-
Polygenic risk score association with multiple sclerosis susceptibility and phenotype in Europeans.Brain. 2023 Feb 13;146(2):645-656. doi: 10.1093/brain/awac092. Brain. 2023. PMID: 35253861 Free PMC article.
-
Genetics and pathogenesis of multiple sclerosis.Semin Immunol. 2009 Dec;21(6):328-33. doi: 10.1016/j.smim.2009.08.003. Epub 2009 Sep 22. Semin Immunol. 2009. PMID: 19775910 Free PMC article. Review.
-
Multiple sclerosis: risk factors, prodromes, and potential causal pathways.Lancet Neurol. 2010 Jul;9(7):727-39. doi: 10.1016/S1474-4422(10)70094-6. Lancet Neurol. 2010. PMID: 20610348 Review.
Cited by
-
Antibody Cross-Reactivity in Auto-Immune Diseases.Int J Mol Sci. 2023 Sep 2;24(17):13609. doi: 10.3390/ijms241713609. Int J Mol Sci. 2023. PMID: 37686415 Free PMC article. Review.
-
Multiple Sclerosis Polygenic Risk Is Not Enriched in Three Multicase Families in Comparison to Population-Based Cases.Hum Mutat. 2024 May 16;2024:9268911. doi: 10.1155/2024/9268911. eCollection 2024. Hum Mutat. 2024. PMID: 40225927 Free PMC article.
-
The immunology of multiple sclerosis.Nat Rev Immunol. 2022 Dec;22(12):734-750. doi: 10.1038/s41577-022-00718-z. Epub 2022 May 4. Nat Rev Immunol. 2022. PMID: 35508809 Review.
-
Age-specific effects of childhood body mass index on multiple sclerosis risk.J Neurol. 2022 Sep;269(9):5052-5060. doi: 10.1007/s00415-022-11161-4. Epub 2022 May 9. J Neurol. 2022. PMID: 35532785 Free PMC article.
-
Prevention of MS Requires Intervention on the Causes of the Disease: Reconciling Genes, Epigenetics, and Epstein Barr Virus.Front Neurol. 2022 Feb 22;13:817677. doi: 10.3389/fneur.2022.817677. eCollection 2022. Front Neurol. 2022. PMID: 35273557 Free PMC article.
References
-
- Olsson T, Barcellos LF, Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol. 2017;13(1):25-36. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
