Prevotella diversity, niches and interactions with the human host
- PMID: 34050328
- PMCID: PMC11290707
- DOI: 10.1038/s41579-021-00559-y
Prevotella diversity, niches and interactions with the human host
Abstract
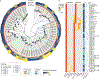
The genus Prevotella includes more than 50 characterized species that occur in varied natural habitats, although most Prevotella spp. are associated with humans. In the human microbiome, Prevotella spp. are highly abundant in various body sites, where they are key players in the balance between health and disease. Host factors related to diet, lifestyle and geography are fundamental in affecting the diversity and prevalence of Prevotella species and strains in the human microbiome. These factors, along with the ecological relationship of Prevotella with other members of the microbiome, likely determine the extent of the contribution of Prevotella to human metabolism and health. Here we review the diversity, prevalence and potential connection of Prevotella spp. in the human host, highlighting how genomic methods and analysis have improved and should further help in framing their ecological role. We also provide suggestions for future research to improve understanding of the possible functions of Prevotella spp. and the effects of the Western lifestyle and diet on the host-Prevotella symbiotic relationship in the context of maintaining human health.
© 2021. Springer Nature Limited.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures


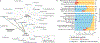
References
-
-
Shah HN & Collins DM Prevotella, a new genus to include Bacteroides melaninogenicus and related species formerly classified in the genus Bacteroides. Int. J. Syst. Bacteriol 40, 205–208 (1990).
This work describes the initial identification, naming and description of the genus Prevotella.
-
-
- Oliver WW & Wherry WB Notes on some bacterial parasites of the human mucous membranes. J. Infect. Dis 28, 341–344 (1921).
-
- Shah HN, Chattaway MA, Rajakurana L. & Gharbia SE Prevotella. Bergey’s Manual of Systematics of Archaea and Bacteria 1–25 (Springer, 2015).
-
-
Fehlner-Peach H. et al. distinct polysaccharide utilization profiles of human intestinal Prevotella copri isolates. Cell Host Microbe 26, 680–690.e5 (2019).
This study highlights how different strains in the P. copri complex have different abilities to target different types of polysaccharides.
-
-
- Gmür R. & Thurnheer T. Direct quantitative differentiation between Prevotella intermedia and Prevotella nigrescens in clinical specimens. Microbiology 148, 1379–1387 (2002). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

