MHC Haplotyping of SARS-CoV-2 Patients: HLA Subtypes Are Not Associated with the Presence and Severity of COVID-19 in the Israeli Population
- PMID: 34050837
- PMCID: PMC8164405
- DOI: 10.1007/s10875-021-01071-x
MHC Haplotyping of SARS-CoV-2 Patients: HLA Subtypes Are Not Associated with the Presence and Severity of COVID-19 in the Israeli Population
Abstract
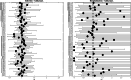
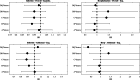
HLA haplotypes were found to be associated with increased risk for viral infections or disease severity in various diseases, including SARS. Several genetic variants are associated with COVID-19 severity. Studies have proposed associations, based on a very small sample and a large number of tested HLA alleles, but no clear association between HLA and COVID-19 incidence or severity has been reported. We conducted a large-scale HLA analysis of Israeli individuals who tested positive for SARS-CoV-2 infection by PCR. Overall, 72,912 individuals with known HLA haplotypes were included in the study, of whom 6413 (8.8%) were found to have SARS-CoV-2 by PCR. A total of 20,937 subjects were of Ashkenazi origin (at least 2/4 grandparents). One hundred eighty-one patients (2.8% of the infected) were hospitalized due to the disease. None of the 66 most common HLA loci (within the five HLA subgroups: A, B, C, DQB1, DRB1) was found to be associated with SARS-CoV-2 infection or hospitalization in the general Israeli population. Similarly, no association was detected in the Ashkenazi Jewish subset. Moreover, no association was found between heterozygosity in any of the HLA loci and either infection or hospitalization. We conclude that HLA haplotypes are not a major risk/protecting factor among the Israeli population for SARS-CoV-2 infection or severity. Our results suggest that if any HLA association exists with the disease it is very weak, and of limited effect on the pandemic.
Keywords: Ashkenazi; HLA; Israel; SARS-CoV-2; association; hospitalization.
© 2021. The Author(s), under exclusive licence to Springer Science+Business Media, LLC, part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
HLA repertoire of 115 UAE nationals infected with SARS-CoV-2.Hum Immunol. 2022 Jan;83(1):1-9. doi: 10.1016/j.humimm.2021.08.012. Epub 2021 Aug 21. Hum Immunol. 2022. PMID: 34462158 Free PMC article.
-
Sex-Specific HLA Alleles Contribute to the Modulation of COVID-19 Severity.Int J Mol Sci. 2024 Dec 8;25(23):13198. doi: 10.3390/ijms252313198. Int J Mol Sci. 2024. PMID: 39684907 Free PMC article.
-
Association between human leukocyte antigen alleles and COVID-19 disease severity.J Infect Public Health. 2024 Sep;17(9):102498. doi: 10.1016/j.jiph.2024.102498. Epub 2024 Jul 15. J Infect Public Health. 2024. PMID: 39173558
-
Association between HLA genotypes and COVID-19 susceptibility, severity and progression: a comprehensive review of the literature.Eur J Med Res. 2021 Aug 3;26(1):84. doi: 10.1186/s40001-021-00563-1. Eur J Med Res. 2021. PMID: 34344463 Free PMC article. Review.
-
Bioinformatic HLA Studies in the Context of SARS-CoV-2 Pandemic and Review on Association of HLA Alleles with Preexisting Medical Conditions.Biomed Res Int. 2021 May 28;2021:6693909. doi: 10.1155/2021/6693909. eCollection 2021. Biomed Res Int. 2021. PMID: 34136572 Free PMC article. Review.
Cited by
-
HLA alleles associated with COVID-19 susceptibility and severity in different populations: a systematic review.Egypt J Med Hum Genet. 2023;24(1):10. doi: 10.1186/s43042-023-00390-5. Epub 2023 Jan 22. Egypt J Med Hum Genet. 2023. PMID: 36710951 Free PMC article. Review.
-
Association between HLA genetics and SARS-CoV-2 infection in a large real-world cohort.Genes Immun. 2025 Jun;26(3):213-221. doi: 10.1038/s41435-025-00328-4. Epub 2025 Apr 24. Genes Immun. 2025. PMID: 40275118 Free PMC article.
-
The Influence of HLA Polymorphisms on the Severity of COVID-19 in the Romanian Population.Int J Mol Sci. 2024 Jan 22;25(2):1326. doi: 10.3390/ijms25021326. Int J Mol Sci. 2024. PMID: 38279325 Free PMC article.
-
The role of HLA genetic variants in COVID-19 susceptibility, severity, and mortality: A global review.J Clin Lab Anal. 2024 Jan;38(1-2):e25005. doi: 10.1002/jcla.25005. Epub 2024 Jan 22. J Clin Lab Anal. 2024. PMID: 38251811 Free PMC article. Review.
-
DOCK2 is involved in the host genetics and biology of severe COVID-19.Nature. 2022 Sep;609(7928):754-760. doi: 10.1038/s41586-022-05163-5. Epub 2022 Aug 8. Nature. 2022. PMID: 35940203 Free PMC article.
References
-
- Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, et al. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–7775. doi: 10.4049/jimmunol.0900111. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

