Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease
- PMID: 34051148
- PMCID: PMC8125900
- DOI: 10.1016/j.immuni.2021.05.010
Longitudinal analysis reveals that delayed bystander CD8+ T cell activation and early immune pathology distinguish severe COVID-19 from mild disease
Abstract
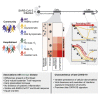
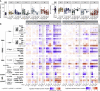
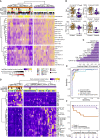
The kinetics of the immune changes in COVID-19 across severity groups have not been rigorously assessed. Using immunophenotyping, RNA sequencing, and serum cytokine analysis, we analyzed serial samples from 207 SARS-CoV2-infected individuals with a range of disease severities over 12 weeks from symptom onset. An early robust bystander CD8+ T cell immune response, without systemic inflammation, characterized asymptomatic or mild disease. Hospitalized individuals had delayed bystander responses and systemic inflammation that was already evident near symptom onset, indicating that immunopathology may be inevitable in some individuals. Viral load did not correlate with this early pathological response but did correlate with subsequent disease severity. Immune recovery is complex, with profound persistent cellular abnormalities in severe disease correlating with altered inflammatory responses, with signatures associated with increased oxidative phosphorylation replacing those driven by cytokines tumor necrosis factor (TNF) and interleukin (IL)-6. These late immunometabolic and immune defects may have clinical implications.
Keywords: COVID-19; SARS-CoV-2; TNF-α; bystander CD8+ T cell; complement; immune pathology; interferon; recovery; systemic inflammation.
Crown Copyright © 2021. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare they have no competing interests. E.J.M. Toonen is an employee of Hycult Biotechnology b.v.
Figures





References
-
- Akbari P., Vuckovic D., Jiang T., Kundu K., Kreuzhuber R., Bao E.L., Mayer L., Collins J.H., Downes K., Georges M. Genetic Analyses of Blood Cell Structure for Biological and Pharmacological Inference. bioRxiv. 2020 doi: 10.1101/2020.01.30.927483. - DOI
-
- Antin J.H., Emerson S.G., Martin P., Gadol N., Ault K.A. Leu-1+ (CD5+) B cells. A major lymphoid subpopulation in human fetal spleen: phenotypic and functional studies. J. Immunol. 1986;136:505–510. - PubMed
-
- Bantug G.R., Galluzzi L., Kroemer G., Hess C. The spectrum of T cell metabolism in health and disease. Nat. Rev. Immunol. 2018;18:19–34. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MC_PC_17230/MRC_/Medical Research Council/United Kingdom
- MR/V011561/1/MRC_/Medical Research Council/United Kingdom
- 210688/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- 209749/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- MR/S036113/1/MRC_/Medical Research Council/United Kingdom
- MR/W014556/1/MRC_/Medical Research Council/United Kingdom
- 207498/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- MR/P008801/1/MRC_/Medical Research Council/United Kingdom
- 200871/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MR/V028448/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00002/10/MRC_/Medical Research Council/United Kingdom
- MR/S00081X/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

