Choice of selectable marker affects recombinant protein expression in cells and exosomes
- PMID: 34051235
- PMCID: PMC8258971
- DOI: 10.1016/j.jbc.2021.100838
Choice of selectable marker affects recombinant protein expression in cells and exosomes
Abstract
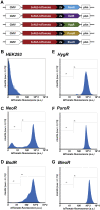
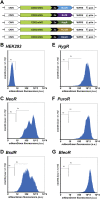
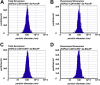
Transgenic mammalian cells are used for numerous research, pharmaceutical, industrial, and clinical purposes, and dominant selectable markers are often used to enable the selection of transgenic cell lines. Using HEK293 cells, we show here that the choice of selectable marker gene has a significant impact on both the level of recombinant protein expression and the cell-to-cell variability in recombinant protein expression. Specifically, we observed that cell lines generated with the NeoR or BsdR selectable markers and selected in the antibiotics G418 or blasticidin, respectively, displayed the lowest level of recombinant protein expression as well as the greatest cell-to-cell variability in transgene expression. In contrast, cell lines generated with the BleoR marker and selected in zeocin yielded cell lines that expressed the highest levels of linked recombinant protein, approximately 10-fold higher than those selected using the NeoR or BsdR markers, as well as the lowest cell-to-cell variability in recombinant protein expression. Intermediate yet still-high levels of expression were observed in cells generated with the PuroR- or HygR-based vectors and that were selected in puromycin or hygromycin, respectively. Similar results were observed in the African green monkey cell line COS7. These data indicate that each combination of selectable marker and antibiotic establishes a threshold below which no cell can survive and that these thresholds vary significantly between different selectable markers. Moreover, we show that choice of selectable marker also affects recombinant protein expression in cell-derived exosomes, consistent with the hypothesis that exosome protein budding is a stochastic rather than determinative process.
Keywords: antibiotics; cell culture; exosomes (vesicles); extracellular vesicle; recombinant protein expression; selectable markers; tetraspanin; transgenic.
Copyright © 2021. Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest F. K. F., C. G., S. J. T, and S. J. G. are coinventors of materials described in this report that are owned by Johns Hopkins University, and if licensed for commercial uses will return royalties to each. S. J. G. currently holds equity in companies that may potentially benefit from the information described in this report, either indirectly or directly.
Figures






Similar articles
-
Degron tagging of BleoR and other antibiotic-resistance genes selects for higher expression of linked transgenes and improved exosome engineering.J Biol Chem. 2022 May;298(5):101846. doi: 10.1016/j.jbc.2022.101846. Epub 2022 Mar 18. J Biol Chem. 2022. PMID: 35314197 Free PMC article.
-
Multiplexed Transgenic Selection and Counterselection Strategies to Expedite Genetic Manipulation Workflows Using Drosophila melanogaster.Curr Protoc. 2023 Feb;3(2):e652. doi: 10.1002/cpz1.652. Curr Protoc. 2023. PMID: 36757287 Free PMC article.
-
Split selectable markers.Nat Commun. 2019 Oct 31;10(1):4968. doi: 10.1038/s41467-019-12891-2. Nat Commun. 2019. PMID: 31672965 Free PMC article.
-
Selectable marker genes in transgenic plants: applications, alternatives and biosafety.J Biotechnol. 2004 Feb 5;107(3):193-232. doi: 10.1016/j.jbiotec.2003.10.011. J Biotechnol. 2004. PMID: 14736458 Review.
-
Selectable marker genes and unintended changes to the plant transcriptome.Plant Biotechnol J. 2009 Apr;7(3):211-8. doi: 10.1111/j.1467-7652.2009.00400.x. Epub 2009 Feb 24. Plant Biotechnol J. 2009. PMID: 19261135 Review.
Cited by
-
Enhanced kinase translocation reporters for simultaneous real-time measurement of PKA, ERK, and calcium.J Biol Chem. 2025 Mar;301(3):108183. doi: 10.1016/j.jbc.2025.108183. Epub 2025 Jan 13. J Biol Chem. 2025. PMID: 39814226 Free PMC article.
-
Exomap1 mouse: A transgenic model for in vivo studies of exosome biology.Extracell Vesicle. 2023 Dec;2:100030. doi: 10.1016/j.vesic.2023.100030. Epub 2023 Oct 10. Extracell Vesicle. 2023. PMID: 39372847 Free PMC article.
-
Endocytosis blocks the vesicular secretion of exosome marker proteins.Sci Adv. 2024 May 10;10(19):eadi9156. doi: 10.1126/sciadv.adi9156. Epub 2024 May 8. Sci Adv. 2024. PMID: 38718108 Free PMC article.
-
Antigen-display exosomes provide adjuvant-free protection against SARS-CoV-2 disease at nanogram levels of spike protein.bioRxiv [Preprint]. 2024 Jan 5:2024.01.04.574272. doi: 10.1101/2024.01.04.574272. bioRxiv. 2024. PMID: 38328234 Free PMC article. Preprint.
-
A Flexible Hybrid Site-Specific Integration-Based Expression System in CHO Cells for Higher-Throughput Evaluation of Monoclonal Antibody Expression Cassettes.Biotechnol J. 2025 Jan;20(1):e202400520. doi: 10.1002/biot.202400520. Biotechnol J. 2025. PMID: 39834086 Free PMC article.
References
-
- Mulligan R.C., Berg P. Expression of a bacterial gene in mammalian cells. Science. 1980;209:1422–1427. - PubMed
-
- Southern P.J., Berg P. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1982;1:327–341. - PubMed
-
- Hunter M., Yuan P., Vavilala D., Fox M. Optimization of protein expression in mammalian cells. Curr. Protoc. Protein Sci. 2019;95:e77. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

