Digital PCR assay for the effective detection of COVID-19 patients with SARS-CoV-2 low viral load
- PMID: 34051244
- PMCID: PMC8149472
- DOI: 10.1016/j.jviromet.2021.114185
Digital PCR assay for the effective detection of COVID-19 patients with SARS-CoV-2 low viral load
Abstract
Objective: Viral nucleic acid detection by real-time reverse transcription polymerase chain reaction (qPCR) is the current standard method for diagnosis of SARS-CoV-2 infection. However, due to low viral load in some COVID-19 patients, false negative results from this method have been repeatedly reported.
Method: In this study, we compared the sensitivity and specificity of digital PCR (dPCR) in simulated samples and clinical samples with qPCR assay through a series of vigorous tests.
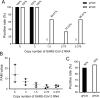
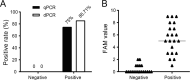
Results: The results showed that dPCR was more sensitive than qPCR especially for samples with low viral load (≤3 copies). In addition, dPCR had similar specificity as qPCR and could effectively distinguish other human coronaviruses and influenza virus from SARS-CoV-2. More importantly, dPCR was more sensitive than qPCR in detecting the virus in the "negative" samples from recurrent COVID-19 patients.
Conclusions: In summary, dPCR could serve as a powerful complement to the current qPCR method for SARS-CoV-2 detection, especially for the samples with extremely low viral load, such as recurrent COVID-19 patients.
Keywords: Digital PCR (dPCR); Recurrent COVID-19 patient; SARS-CoV-2; Sensitivity; Specificity.
Copyright © 2021. Published by Elsevier B.V.
Conflict of interest statement
The authors have no conflicts of interests to declare.
Figures


Similar articles
-
Digital PCR Applications in the SARS-CoV-2/COVID-19 Era: a Roadmap for Future Outbreaks.Clin Microbiol Rev. 2022 Sep 21;35(3):e0016821. doi: 10.1128/cmr.00168-21. Epub 2022 Mar 8. Clin Microbiol Rev. 2022. PMID: 35258315 Free PMC article. Review.
-
The potential application of digital PCR in detecting different SARS-CoV-2 viral loads.J Virol Methods. 2025 Jun;335:115151. doi: 10.1016/j.jviromet.2025.115151. Epub 2025 Apr 1. J Virol Methods. 2025. PMID: 40180223
-
Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR.Talanta. 2021 Mar 1;224:121726. doi: 10.1016/j.talanta.2020.121726. Epub 2020 Oct 27. Talanta. 2021. PMID: 33379001 Free PMC article.
-
Detection of SARS-CoV-2 RNA in nasopharyngeal swabs from COVID-19 patients and asymptomatic cases of infection by real-time and digital PCR.Klin Lab Diagn. 2020 Dec 29;65(12):785-792. doi: 10.18821/0869-2084-2020-65-12-785-792. Klin Lab Diagn. 2020. PMID: 33373511 English.
-
Diagnostic performances of common nucleic acid tests for SARS-CoV-2 in hospitals and clinics: a systematic review and meta-analysis.Lancet Microbe. 2021 Dec;2(12):e704-e714. doi: 10.1016/S2666-5247(21)00214-7. Epub 2021 Oct 13. Lancet Microbe. 2021. PMID: 34661181 Free PMC article.
Cited by
-
High concordance in SARSCoV-2 detection between automated (Abbott m2000) and manual (DaAn gene) RT-PCR systems: The EDCTP PERFECT-Study in Cameroon.J Public Health Afr. 2022 May 24;13(1):2163. doi: 10.4081/jphia.2022.2163. eCollection 2022 May 24. J Public Health Afr. 2022. PMID: 35720798 Free PMC article.
-
Digital PCR (dPCR) Quantification of miR-155-5p as a Potential Candidate for a Tissue Biomarker of Inflammation in Rabbits Infected with Lagovirus europaeus/Rabbit Hemorrhagic Disease Virus (RHDV).Viruses. 2023 Jul 19;15(7):1578. doi: 10.3390/v15071578. Viruses. 2023. PMID: 37515264 Free PMC article.
-
Digital PCR Applications in the SARS-CoV-2/COVID-19 Era: a Roadmap for Future Outbreaks.Clin Microbiol Rev. 2022 Sep 21;35(3):e0016821. doi: 10.1128/cmr.00168-21. Epub 2022 Mar 8. Clin Microbiol Rev. 2022. PMID: 35258315 Free PMC article. Review.
-
Droplet Digital PCR or Real-Time PCR as a Method for Quantifying SARS-CoV-2 RNA in Plasma-Is There a Difference?Viruses. 2025 May 28;17(6):772. doi: 10.3390/v17060772. Viruses. 2025. PMID: 40573363 Free PMC article.
-
New RT-PCR Assay for the Detection of Current and Future SARS-CoV-2 Variants.Viruses. 2023 Jan 11;15(1):206. doi: 10.3390/v15010206. Viruses. 2023. PMID: 36680246 Free PMC article.
References
-
- Alteri C., Cento V., Antonello M., Colagrossi L., Merli M., Ughi N., Renica S., Matarazzo E., Di Ruscio F., Tartaglione L., et al. Detection and quantification of SARS-CoV-2 by droplet digital PCR in real-time PCR negative nasopharyngeal swabs from suspected COVID-19 patients. PLoS One. 2020;15(9):10. - PMC - PubMed
-
- Basu A.S. Digital assays part I: partitioning statistics and digital PCR. SLAS Technol. 2017;22(4):369–386. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

