Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance
- PMID: 34051492
- PMCID: PMC8542657
- DOI: 10.1016/j.scitotenv.2021.147829
Feasibility of neighborhood and building scale wastewater-based genomic epidemiology for pathogen surveillance
Abstract
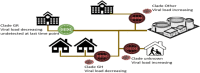
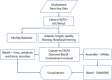
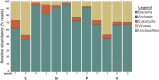
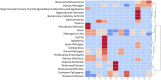
The benefits of wastewater-based epidemiology (WBE) for tracking the viral load of SARS-CoV-2, the causative agent of COVID-19, have become apparent since the start of the pandemic. However, most sampling occurs at the wastewater treatment plant influent and therefore monitors the entire catchment, encompassing multiple municipalities, and is conducted using quantitative polymerase chain reaction (qPCR), which only quantifies one target. Sequencing methods provide additional strain information and also can identify other pathogens, broadening the applicability of WBE to beyond the COVID-19 pandemic. Here we demonstrate feasibility of sampling at the neighborhood or building complex level using qPCR, targeted sequencing, and untargeted metatranscriptomics (total RNA sequencing) to provide a refined understanding of the local dynamics of SARS-CoV-2 strains and identify other pathogens circulating in the community. We demonstrate feasibility of tracking SARS-CoV-2 at the neighborhood, hospital, and nursing home level with the ability to detect one COVID-19 positive out of 60 nursing home residents. The viral load obtained was correlative with the number of COVID-19 patients being treated in the hospital. Targeted wastewater-based sequencing over time demonstrated that nonsynonymous mutations fluctuate in the viral population. Clades and shifts in mutation profiles within the community were monitored and could be used to determine if vaccine or diagnostics need to be adapted to ensure continued efficacy. Furthermore, untargeted RNA sequencing identified several other pathogens in the samples. Therefore, untargeted RNA sequencing could be used to identify new outbreaks or emerging pathogens beyond the COVID-19 pandemic.
Keywords: Mutation analysis; RNAseq; SARS-CoV-2; Targeted genome sequencing; Viral load; Wastewater based epidemiology.
Copyright © 2021 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., et al. Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54:7754–7757. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

