Dysregulation of COVID-19 related gene expression in the COPD lung
- PMID: 34051791
- PMCID: PMC8164067
- DOI: 10.1186/s12931-021-01755-3
Dysregulation of COVID-19 related gene expression in the COPD lung
Abstract
Background: Chronic obstructive pulmonary disease (COPD) patients are at increased risk of poor outcome from Coronavirus disease (COVID-19). Early data suggest elevated Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) receptor angiotensin converting enzyme 2 (ACE2) expression, but relationships to disease phenotype and downstream regulators of inflammation in the Renin-Angiotensin system (RAS) are unknown. We aimed to determine the relationship between RAS gene expression relevant to SARS-CoV-2 infection in the lung with disease characteristics in COPD, and the regulation of newly identified SARS-CoV-2 receptors and spike-cleaving proteases, important for SARS-CoV-2 infection.
Methods: We quantified gene expression using RNA sequencing of epithelial brushings and bronchial biopsies from 31 COPD and 37 control subjects.
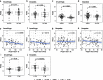
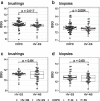
Results: ACE2 gene expression (log2-fold change (FC)) was increased in COPD compared to ex-smoking (HV-ES) controls in epithelial brushings (0.25, p = 0.042) and bronchial biopsies (0.23, p = 0.050), and correlated with worse lung function (r = - 0.28, p = 0.0090). ACE2 was further increased in frequent exacerbators compared to infrequent exacerbators (0.51, p = 0.00045) and associated with use of ACE inhibitors (ACEi) (0.50, p = 0.0034), having cardiovascular disease (0.23, p = 0.048) or hypertension (0.34, p = 0.0089), and inhaled corticosteroid use in COPD subjects in bronchial biopsies (0.33, p = 0.049). Angiotensin II receptor type (AGTR)1 and 2 expression was decreased in COPD bronchial biopsies compared to HV-ES controls with log2FC of -0.26 (p = 0.033) and - 0.40, (p = 0.0010), respectively. However, the AGTR1:2 ratio was increased in COPD subjects compared with HV-ES controls, log2FC of 0.57 (p = 0.0051). Basigin, a newly identified potential SARS-CoV-2 receptor was also upregulated in both brushes, log2FC of 0.17 (p = 0.0040), and bronchial biopsies, (log2FC of 0.18 (p = 0.017), in COPD vs HV-ES. Transmembrane protease, serine (TMPRSS)2 was not differentially regulated between control and COPD. However, various other spike-cleaving proteases were, including TMPRSS4 and Cathepsin B, in both epithelial brushes (log2FC of 0.25 (p = 0.0012) and log2FC of 0.56 (p = 5.49E-06), respectively) and bronchial biopsies (log2FC of 0.49 (p = 0.00021) and log2FC of 0.246 (p = 0.028), respectively).
Conclusion: This study identifies key differences in expression of genes related to susceptibility and aetiology of COVID-19 within the COPD lung. Further studies to understand the impact on clinical course of disease are now required.
Keywords: ACE2; COPD; COVID-19; Infection; Inflammation; SARS-CoV-2.
Conflict of interest statement
Dr. Öberg, Dr. Angermann, Dr. Hühn, Dr. Muthas, Dr. Etal, Dr Belfield, Dr. Karlsson, Dr. Nordström and Dr Ostridge are paid employees of AstraZeneca; Dr. Staples reports grants from AstraZeneca, during the conduct of the study; Dr. Wilkinson reports grants and personal fees from AstraZeneca, during the conduct of the study; personal fees and other from MMH, grants and personal fees from GSK, grants and personal fees from AZ, personal fees from BI, grants and personal fees from Synairgen, outside the submitted work; Dr Watson, Dr. Spalluto, Dr. Burke, Dr. Cellura and Dr. Freeman report no conflicts of interest
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

