Motifier: An IgOme Profiler Based on Peptide Motifs Using Machine Learning
- PMID: 34052285
- PMCID: PMC8286340
- DOI: 10.1016/j.jmb.2021.167071
Motifier: An IgOme Profiler Based on Peptide Motifs Using Machine Learning
Abstract
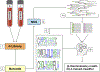
Antibodies provide a comprehensive record of the encounters with threats and insults to the immune system. The ability to examine the repertoire of antibodies in serum and discover those that best represent "discriminating features" characteristic of various clinical situations, is potentially very useful. Recently, phage display technologies combined with Next-Generation Sequencing (NGS) produced a powerful experimental methodology, coined "Deep-Panning", in which the spectrum of serum antibodies is probed. In order to extract meaningful biological insights from the tens of millions of affinity-selected peptides generated by Deep-Panning, advanced bioinformatics algorithms are a must. In this study, we describe Motifier, a computational pipeline comprised of a set of algorithms that systematically generates discriminatory peptide motifs based on the affinity-selected peptides identified by Deep-Panning. These motifs are shown to effectively characterize antibody binding activities and through the implementation of machine-learning protocols are shown to accurately classify complex antibody mixtures representing various biological conditions.
Keywords: deep-panning; next-generation phage display; phage display; random peptide libraries.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures







References
-
- Smith GP, Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface., Science. 228 (1985) 1315–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

