Development and Application of a Fast Method to Acquire the Accurate Whole-Genome Sequences of Human Adenoviruses
- PMID: 34054762
- PMCID: PMC8160523
- DOI: 10.3389/fmicb.2021.661382
Development and Application of a Fast Method to Acquire the Accurate Whole-Genome Sequences of Human Adenoviruses
Abstract
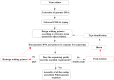
The whole-genome sequencing (WGS) of human adenoviruses (HAdVs) plays an important role in identifying, typing, and mutation analysis of HAdVs. Nowadays, three generations of sequencing have been developed. The accuracy of first-generation sequencing is up to 99.99%, whereas this technology relies on PCR and is time consuming; the next-generation sequencing (NGS) is expensive and not cost effective for determining a few special samples; and the third-generation sequencing technology has a higher error rate. In this study, first, we developed an efficient HAdV genomic DNA extraction method. Using the complete genomic DNA instead of the PCR amplicons as the direct sequencing template and a set of walking primers, we developed the HAdV WGS method based on first-generation sequencing. The HAdV whole genomes were effectively sequenced by a set of one-way sequencing primers designed, which reduced the sequencing time and cost. More importantly, high sequence accuracy is guaranteed. Four HAdV strains (GZ01, GZ02, HK35, and HK91) were isolated from children with acute respiratory diseases (ARDs), and the complete genomes were sequenced using this method. The accurate sequences of the whole inverted terminal repeats (ITRs) at both ends of the HAdV genomes were also acquired. The genome sequence of human adenovirus type 14 (HAdV-B14) strain GZ01 acquired by this method is identical to the sequence released in GenBank, which indicates that this novel sequencing method has high accuracy. The comparative genomic analysis identified that strain GZ02 isolated in September 2010 had the identical genomic sequence with the HAdV-B14 strain GZ01 (October 2010). Therefore, strain GZ02 is the first HAdV-B14 isolate emergent in China (September 2010; GenBank acc no. MW692349). The WGS of HAdV-C2 strain HK91 and HAdV-E4 strain HK35 isolated from children with acute respiratory disease in Hong Kong were also determined by this sequencing method. In conclusion, this WGS method is fast, accurate, and universal for common human adenovirus species B, C, and E. The sequencing strategy may also be applied to the WGS of the other DNA viruses.
Keywords: Sanger sequencing; genomic DNA; human adenovirus; human adenovirus type 14; human adenovirus type 2; human adenovirus type 4; inverted terminal repeats (ITRs); whole-genome sequencing.
Copyright © 2021 Zhao, Guan, Ma, Yan, Ou, Zhang, Yu, Wu and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Centers for Disease Control and Prevention [CDC] (2007). Acute respiratory disease associated with adenovirus serotype 14-four states, 2006-2007. MMWR Morb. Mortal. Wkly. Rep. 56 1181–1184. - PubMed
-
- Cheng Z., Yan Y., Jing S., Li W.-G., Chen W.-W., Zhang J., et al. (2018). Comparative genomic analysis of re-emergent human adenovirus Type 55 pathogens associated with adult severe community-acquired pneumonia reveals conserved genomes and capsid proteins. Front. Microbiol. 9:1180. 10.3389/fmicb.2018.01180 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

