Novel Identification of Bacterial Epigenetic Regulations Would Benefit From a Better Exploitation of Methylomic Data
- PMID: 34054792
- PMCID: PMC8160106
- DOI: 10.3389/fmicb.2021.685670
Novel Identification of Bacterial Epigenetic Regulations Would Benefit From a Better Exploitation of Methylomic Data
Abstract
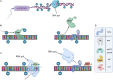
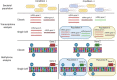
DNA methylation can be part of epigenetic mechanisms, leading to cellular subpopulations with heterogeneous phenotypes. While prokaryotic phenotypic heterogeneity is of critical importance for a successful infection by several major pathogens, the exact mechanisms involved in this phenomenon remain unknown in many cases. Powerful sequencing tools have been developed to allow the detection of the DNA methylated bases at the genome level, and they have recently been extensively applied on numerous bacterial species. Some of these tools are increasingly used for metagenomics analysis but only a limited amount of the available methylomic data is currently being exploited. Because newly developed tools now allow the detection of subpopulations differing in their genome methylation patterns, it is time to emphasize future strategies based on a more extensive use of methylomic data. This will ultimately help to discover new epigenetic gene regulations involved in bacterial phenotypic heterogeneity, including during host-pathogen interactions.
Keywords: DNA methylation; bacterial epigenetics; methylome; phenotypic heterogeneity; transcriptome.
Copyright © 2021 Payelleville and Brillard.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Atack J. M., Guo C., Litfin T., Yang L., Blackall P. J., Zhou Y., et al. (2020a). Systematic analysis of REBASE identifies numerous Type I restriction-modification systems with duplicated, distinct hsdS specificity genes that can switch system specificity by recombination. mSystems 5 e00497–e00520. - PMC - PubMed
-
- Atack J. M., Guo C., Yang L., Zhou Y., Jennings M. P. (2020b). DNA sequence repeats identify numerous Type I restriction-modification systems that are potential epigenetic regulators controlling phase-variable regulons; phasevarions. FASEB J.: Off. Publ. Federation Am. Societies Exp. Biol. 34 1038–1051. 10.1096/fj.201901536rr - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

