Deciphering the Complexity of 3D Chromatin Organization Driving Lymphopoiesis and Lymphoid Malignancies
- PMID: 34054841
- PMCID: PMC8160312
- DOI: 10.3389/fimmu.2021.669881
Deciphering the Complexity of 3D Chromatin Organization Driving Lymphopoiesis and Lymphoid Malignancies
Abstract
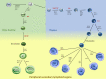
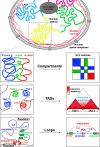
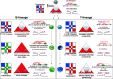
Proper lymphopoiesis and immune responses depend on the spatiotemporal control of multiple processes, including gene expression, DNA recombination and cell fate decisions. High-order 3D chromatin organization is increasingly appreciated as an important regulator of these processes and dysregulation of genomic architecture has been linked to various immune disorders, including lymphoid malignancies. In this review, we present the general principles of the 3D chromatin topology and its dynamic reorganization during various steps of B and T lymphocyte development and activation. We also discuss functional interconnections between architectural, epigenetic and transcriptional changes and introduce major key players of genomic organization in B/T lymphocytes. Finally, we present how alterations in architectural factors and/or 3D genome organization are linked to dysregulation of the lymphopoietic transcriptional program and ultimately to hematological malignancies.
Keywords: 3D chromatin organization; B cells; T cells; activation; lymphoid malignancies; lymphopoiesis.
Copyright © 2021 Scourzic, Salataj and Apostolou.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

