Genome-Wide Association Study for Grain Micronutrient Concentrations in Wheat Advanced Lines Derived From Wild Emmer
- PMID: 34054897
- PMCID: PMC8160437
- DOI: 10.3389/fpls.2021.651283
Genome-Wide Association Study for Grain Micronutrient Concentrations in Wheat Advanced Lines Derived From Wild Emmer
Abstract
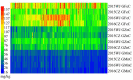
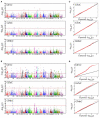
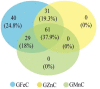
Wheat is one of the important staple crops as the resources of both food and micronutrient for most people of the world. However, the levels of micronutrients (especially Fe and Zn) in common wheat are inherently low. Biofortification is an effective way to increase the micronutrient concentration of wheat. Wild emmer wheat (Triticum turgidum ssp. dicoccoides, AABB, 2n = 4x = 28) is an important germplasm resource for wheat micronutrients improvement. In the present study, a genome-wide association study (GWAS) was performed to characterize grain iron, zinc, and manganese concentration (GFeC, GZnC, and GMnC) in 161 advanced lines derived from wild emmer. Using both the general linear model and mixed linear model, we identified 14 high-confidence significant marker-trait associations (MTAs) that were associated with GFeC, GZnC, and GMnC of which nine MTAs were novel. Six MTAs distributed on chromosomes 3B, 4A, 4B, 5A, and 7B were significantly associated with GFeC. Three MTAs on 1A and 2A were significantly associated with GZnC and five MTAs on 1B were significantly associated with GMnC. These MTAs show no negative effects on thousand kernel weight (TKW), implying the potential value for simultaneous improvement of micronutrient concentrations and TKW in breeding. Meanwhile, the GFeC, GZnC and GMnC are positively correlated, suggesting that these traits could be simultaneously improved. Genotypes containing high-confidence MTAs and 61 top genotypes with a higher concentration of grain micronutrients were recommended for wheat biofortification breeding. A total of 38 candidate genes related to micronutrient concentrations were identified. These candidates can be classified into four main groups: enzymes, transporter proteins, MYB transcription factor, and plant defense responses proteins. The MTAs and associated candidate genes provide essential information for wheat biofortification breeding through marker-assisted selection (MAS).
Keywords: GWAS; biofortification; common wheat; grain micronutrient concentrations; wide hybridization; wild emmer.
Copyright © 2021 Liu, Huang, Li, Liu, Yan, Tang, Zheng, Liu and Wu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer VG declared a past collaboration with two of the authors JL and BW to the handling editor.
Figures

References
-
- Alvarado G., López M., Vargas M., Pacheco Á., Rodríguez F., Burgueño J., et al. (2015). META-R (Multi Environment Trail Analysis with R for Windows) Version 5.0. Texcoco: International Maize and Wheat Improvement Center.
-
- Ates D., Asciogul T. K., Nemli S., Erdogmus S., Esiyok D., Tanyolac M. B. (2018). Association mapping of days to flowering in common bean (Phaseolus vulgaris L.) revealed by DArT markers. Mol. Breed. 38:113. 10.1007/s11032-018-0868-0 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

