GhGPAT12/ 25 Are Essential for the Formation of Anther Cuticle and Pollen Exine in Cotton (Gossypium hirsutum L.)
- PMID: 34054906
- PMCID: PMC8155372
- DOI: 10.3389/fpls.2021.667739
GhGPAT12/ 25 Are Essential for the Formation of Anther Cuticle and Pollen Exine in Cotton (Gossypium hirsutum L.)
Abstract
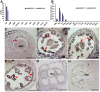
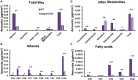
Glycerol-3-phosphate acyltransferases (GPATs), critical for multiple biological processes like male fertility, have been extensively characterized. However, their precise functions and underlying regulatory mechanism in cotton anther development are unclear. This research demonstrated the importance of GhGPAT12/25 (a paralogs pair on A12/D12 sub-chromosome of cotton) to regulate the degradation of tapetum, anther cuticle formation, and pollen exine development. GhGPAT12 and GhGPAT25 exhibited specifically detected transcripts in tapetum and pollen exine during the early anther developmental stages. GhGPAT12/25 are sn-2 glycerol-3-phosphate acyltransferases and can transfer the acyl group of palmitoyl-CoA to glycerol-3-phosphate (G3P). CRISPR/Cas9-mediated knockout identified the functional redundancy of GhGPAT12 and GhGPAT25. Knockout of both genes caused completely male sterility associated with abnormal anther cuticle, swollen tapetum, and inviable microspores with defective exine and irregular unrestricted shape. RNA-seq analysis showed that the loss of function of GhGPAT12/25 affects the processes of wax metabolic, glycerol monomer biosynthesis, and transport. Consistently, cuticular waxes were dramatically reduced in mutant anthers. Yeast one-hybrid system (Y1H), virus-induced gene silencing (VIGS), and dual-luciferase (LUC) assays illustrated that GhMYB80s are likely to directly activate the expression of GhGPAT12/25. This study provides important insights for revealing the regulatory mechanism underlying anther development in cotton.
Keywords: CRISPR/Cas9; GhGPAT12/25; anther cuticle; cotton; male sterility; pollen exine.
Copyright © 2021 Zhang, Wei, Hao, Wu, Ma, Zhang, Wang, Fu, Ma, Lu and Yu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








Similar articles
-
The ZmMYB84-ZmPKSB regulatory module controls male fertility through modulating anther cuticle-pollen exine trade-off in maize anthers.Plant Biotechnol J. 2022 Dec;20(12):2342-2356. doi: 10.1111/pbi.13911. Epub 2022 Sep 7. Plant Biotechnol J. 2022. PMID: 36070225 Free PMC article.
-
ABNORMAL POLLEN VACUOLATION1 (APV1) is required for male fertility by contributing to anther cuticle and pollen exine formation in maize.Plant J. 2017 Apr;90(1):96-110. doi: 10.1111/tpj.13476. Epub 2017 Mar 8. Plant J. 2017. PMID: 28078801
-
IRREGULAR POLLEN EXINE1 Is a Novel Factor in Anther Cuticle and Pollen Exine Formation.Plant Physiol. 2017 Jan;173(1):307-325. doi: 10.1104/pp.16.00629. Epub 2016 Nov 15. Plant Physiol. 2017. PMID: 28049856 Free PMC article.
-
Triphasic regulation of ZmMs13 encoding an ABCG transporter is sequentially required for callose dissolution, pollen exine and anther cuticle formation in maize.J Adv Res. 2023 Jul;49:15-30. doi: 10.1016/j.jare.2022.09.006. Epub 2022 Sep 18. J Adv Res. 2023. PMID: 36130683 Free PMC article.
-
CRISPR/Cas9-mediated knockout of GhAMS11 and GhMS188 reveals key roles in tapetal development and pollen exine formation in upland cotton.Int J Biol Macromol. 2025 Mar;293:139362. doi: 10.1016/j.ijbiomac.2024.139362. Epub 2024 Dec 30. Int J Biol Macromol. 2025. PMID: 39743080
Cited by
-
Genome-Wide Identification, Characterization, Evolutionary Analysis, and Expression Pattern of the GPAT Gene Family in Barley and Functional Analysis of HvGPAT18 under Abiotic Stress.Int J Mol Sci. 2024 Jun 1;25(11):6101. doi: 10.3390/ijms25116101. Int J Mol Sci. 2024. PMID: 38892304 Free PMC article.
-
High-temperature stress in crops: male sterility, yield loss and potential remedy approaches.Plant Biotechnol J. 2023 Apr;21(4):680-697. doi: 10.1111/pbi.13946. Epub 2022 Nov 4. Plant Biotechnol J. 2023. PMID: 36221230 Free PMC article. Review.
-
Applications of Virus-Induced Gene Silencing in Cotton.Plants (Basel). 2024 Jan 17;13(2):272. doi: 10.3390/plants13020272. Plants (Basel). 2024. PMID: 38256825 Free PMC article. Review.
-
Functional redundancy of transcription factors SlNOR and SlNOR-like1 is required for pollen development in tomato.Hortic Res. 2025 Jan 6;12(4):uhaf003. doi: 10.1093/hr/uhaf003. eCollection 2025 Apr. Hortic Res. 2025. PMID: 40078722 Free PMC article.
-
Use of CRISPR/Cas9-Based Gene Editing to Simultaneously Mutate Multiple Homologous Genes Required for Pollen Development and Male Fertility in Maize.Cells. 2022 Jan 27;11(3):439. doi: 10.3390/cells11030439. Cells. 2022. PMID: 35159251 Free PMC article.
References
-
- Ariizumi T., Hatakeyama K., Hinata K., Sato S., Kato T., Tabata S., et al. (2003). A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 53 107–116. 10.1023/b:plan.0000009269.97773.70 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

