Identification of Potential Pathogenic Super-Enhancers-Driven Genes in Pulmonary Fibrosis
- PMID: 34054916
- PMCID: PMC8153712
- DOI: 10.3389/fgene.2021.644143
Identification of Potential Pathogenic Super-Enhancers-Driven Genes in Pulmonary Fibrosis
Abstract
Abnormal fibroblast differentiation into myofibroblast is a crucial pathological mechanism of pulmonary fibrosis (PF). Super-enhancers, a newly discovered cluster of regulatory elements, are regarded as the regulators of cell identity. We speculate that abnormal activation of super-enhancers must be involved in the pathological process of PF. This study aims to identify potential pathogenic super-enhancer-driven genes in PF. Differentially expressed genes (DEGs) in PF mouse lungs were identified from a GEO dataset (GDS1492). We collected super-enhancers and their associated genes in human lung fibroblasts and mouse embryonic fibroblasts from SEA version 3.0, a network database that provides comprehensive information on super-enhancers. We crosslinked upregulated DEGs and super-enhancer-associated genes in fibroblasts to predict potential super-enhancer-driven pathogenic genes in PF. A total of 25 genes formed an overlap, and the protein-protein interaction network of these genes was constructed by the STRING database. An interaction network of transcription factors (TFs), super-enhancers, and associated genes was constructed using the Cytoscape software. Gene enrichment analyses, including KEGG pathway and GO analysis, were performed for these genes. Latent transforming growth factor beta (TGF-β) binding protein 2 (LTBP2), one of the predicted super-enhancer-driven pathogenic genes, was used to verify the predicted network's accuracy. LTBP2 was upregulated in the lungs of the bleomycin-induced PF mouse model and TGF-β1-stimulated mouse and human fibroblasts. Myc is one of the TFs binding to the LTBP2 super-enhancer. Knockout of super-enhancer sequences with a CRISPR/Cas9 plasmid or inhibition of Myc all decreased TGF-β1-induced LTBP2 expression in NIH/3 T3 cells. Identifying and interfering super-enhancers might be a new way to explore possible therapeutic methods for PF.
Keywords: LTBP2; fibroblast; pulmonary fibrosis; super-enhancer; transcription factor.
Copyright © 2021 Li, Zhao, Li, Yao, Zhang, Si, Liu, Jiang and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
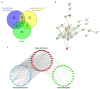



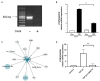
Similar articles
-
Latent Transforming Growth Factor-β Binding Protein-2 Regulates Lung Fibroblast-to-Myofibroblast Differentiation in Pulmonary Fibrosis via NF-κB Signaling.Front Pharmacol. 2021 Dec 24;12:788714. doi: 10.3389/fphar.2021.788714. eCollection 2021. Front Pharmacol. 2021. PMID: 35002722 Free PMC article.
-
LTBP2 is secreted from lung myofibroblasts and is a potential biomarker for idiopathic pulmonary fibrosis.Clin Sci (Lond). 2018 Jul 31;132(14):1565-1580. doi: 10.1042/CS20180435. Print 2018 Jul 31. Clin Sci (Lond). 2018. PMID: 30006483 Free PMC article.
-
SEA: a super-enhancer archive.Nucleic Acids Res. 2016 Jan 4;44(D1):D172-9. doi: 10.1093/nar/gkv1243. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26578594 Free PMC article.
-
Targeting super-enhancer activity for colorectal cancer therapy.Am J Transl Res. 2024 Mar 15;16(3):700-719. doi: 10.62347/QKHB5897. eCollection 2024. Am J Transl Res. 2024. PMID: 38586095 Free PMC article. Review.
-
Super-enhancers in cancer.Pharmacol Ther. 2019 Jul;199:129-138. doi: 10.1016/j.pharmthera.2019.02.014. Epub 2019 Mar 16. Pharmacol Ther. 2019. PMID: 30885876 Review.
Cited by
-
Analysis of super-enhancer using machine learning and its application to medical biology.Brief Bioinform. 2023 May 19;24(3):bbad107. doi: 10.1093/bib/bbad107. Brief Bioinform. 2023. PMID: 36960780 Free PMC article. Review.
-
Differential Expression of Super-Enhancer-Associated Long Non-coding RNAs in Uterine Leiomyomas.Reprod Sci. 2022 Oct;29(10):2960-2976. doi: 10.1007/s43032-022-00981-4. Epub 2022 May 31. Reprod Sci. 2022. PMID: 35641855 Free PMC article.
-
Nintedanib Alleviates Experimental Colitis by Inhibiting CEBPB/PCK1 and CEBPB/EFNA1 Pathways.Front Pharmacol. 2022 Jul 14;13:904420. doi: 10.3389/fphar.2022.904420. eCollection 2022. Front Pharmacol. 2022. PMID: 35910380 Free PMC article.
-
Latent Transforming Growth Factor-β Binding Protein-2 Regulates Lung Fibroblast-to-Myofibroblast Differentiation in Pulmonary Fibrosis via NF-κB Signaling.Front Pharmacol. 2021 Dec 24;12:788714. doi: 10.3389/fphar.2021.788714. eCollection 2021. Front Pharmacol. 2021. PMID: 35002722 Free PMC article.
-
Anti-Inflammatory and Anti-Fibrotic Effect of Immortalized Mesenchymal-Stem-Cell-Derived Conditioned Medium on Human Lung Myofibroblasts and Epithelial Cells.Int J Mol Sci. 2022 Apr 20;23(9):4570. doi: 10.3390/ijms23094570. Int J Mol Sci. 2022. PMID: 35562961 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

