Prediction of repurposed drugs for Coronaviruses using artificial intelligence and machine learning
- PMID: 34055238
- PMCID: PMC8141697
- DOI: 10.1016/j.csbj.2021.05.037
Prediction of repurposed drugs for Coronaviruses using artificial intelligence and machine learning
Abstract
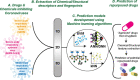
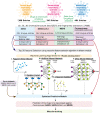
The world is facing the COVID-19 pandemic caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Likewise, other viruses of the Coronaviridae family were responsible for causing epidemics earlier. To tackle these viruses, there is a lack of approved antiviral drugs. Therefore, we have developed robust computational methods to predict the repurposed drugs using machine learning techniques namely Support Vector Machine, Random Forest, k-Nearest Neighbour, Artificial Neural Network, and Deep Learning. We used the experimentally validated drugs/chemicals with anticorona activity (IC50/EC50) from 'DrugRepV' repository. The unique entries of SARS-CoV-2 (142), SARS (221), MERS (123), and overall Coronaviruses (414) were subdivided into the training/testing and independent validation datasets, followed by the extraction of chemical/structural descriptors and fingerprints (17968). The highly relevant features were filtered using the recursive feature selection algorithm. The selected chemical descriptors were used to develop prediction models with Pearson's correlation coefficients ranging from 0.60 to 0.90 on training/testing. The robustness of the predictive models was further ensured using external independent validation datasets, decoy datasets, applicability domain, and chemical analyses. The developed models were used to predict promising repurposed drug candidates against coronaviruses after scanning the DrugBank. Top predicted molecules for SARS-CoV-2 were further validated by molecular docking against the spike protein complex with ACE receptor. We found potential repurposed drugs namely Verteporfin, Alatrofloxacin, Metergoline, Rescinnamine, Leuprolide, and Telotristat ethyl with high binding affinity. These 'anticorona' computational models would assist in antiviral drug discovery against SARS-CoV-2 and other Coronaviruses.
Keywords: AI; COVID-19; Chemical descriptors; Coronaviruses; Drug repurposing; Machine learning; SARS-CoV-2.
© 2021 The Authors. Published by Elsevier B.V. on behalf of Research Network of Computational and Structural Biotechnology.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





Similar articles
-
Targeting non-structural proteins of Hepatitis C virus for predicting repurposed drugs using QSAR and machine learning approaches.Comput Struct Biotechnol J. 2022 Jun 30;20:3422-3438. doi: 10.1016/j.csbj.2022.06.060. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35832613 Free PMC article.
-
Exploiting cheminformatic and machine learning to navigate the available chemical space of potential small molecule inhibitors of SARS-CoV-2.Comput Struct Biotechnol J. 2021;19:424-438. doi: 10.1016/j.csbj.2020.12.028. Epub 2020 Dec 29. Comput Struct Biotechnol J. 2021. PMID: 33391634 Free PMC article.
-
DrugRepV: a compendium of repurposed drugs and chemicals targeting epidemic and pandemic viruses.Brief Bioinform. 2021 Mar 22;22(2):1076-1084. doi: 10.1093/bib/bbaa421. Brief Bioinform. 2021. PMID: 33480398 Free PMC article.
-
Current Strategies of Antiviral Drug Discovery for COVID-19.Front Mol Biosci. 2021 May 13;8:671263. doi: 10.3389/fmolb.2021.671263. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055887 Free PMC article. Review.
-
Current status of antivirals and druggable targets of SARS CoV-2 and other human pathogenic coronaviruses.Drug Resist Updat. 2020 Dec;53:100721. doi: 10.1016/j.drup.2020.100721. Epub 2020 Aug 26. Drug Resist Updat. 2020. PMID: 33132205 Free PMC article. Review.
Cited by
-
Biofilm-i: A Platform for Predicting Biofilm Inhibitors Using Quantitative Structure-Relationship (QSAR) Based Regression Models to Curb Antibiotic Resistance.Molecules. 2022 Jul 29;27(15):4861. doi: 10.3390/molecules27154861. Molecules. 2022. PMID: 35956807 Free PMC article.
-
SperoPredictor: An Integrated Machine Learning and Molecular Docking-Based Drug Repurposing Framework With Use Case of COVID-19.Front Public Health. 2022 Jun 16;10:902123. doi: 10.3389/fpubh.2022.902123. eCollection 2022. Front Public Health. 2022. PMID: 35784208 Free PMC article.
-
A review of SARS-CoV-2 drug repurposing: databases and machine learning models.Front Pharmacol. 2023 Aug 4;14:1182465. doi: 10.3389/fphar.2023.1182465. eCollection 2023. Front Pharmacol. 2023. PMID: 37601065 Free PMC article. Review.
-
Computational identification of repurposed drugs against viruses causing epidemics and pandemics via drug-target network analysis.Comput Biol Med. 2021 Sep;136:104677. doi: 10.1016/j.compbiomed.2021.104677. Epub 2021 Jul 23. Comput Biol Med. 2021. PMID: 34332351 Free PMC article.
-
Anti-Dengue: A Machine Learning-Assisted Prediction of Small Molecule Antivirals against Dengue Virus and Implications in Drug Repurposing.Viruses. 2023 Dec 27;16(1):45. doi: 10.3390/v16010045. Viruses. 2023. PMID: 38257744 Free PMC article.
References
-
- Woo P.C.Y., Lau S.K.P., Lam C.S.F., Lau C.C.Y., Tsang A.K.L., Lau J.H.N. Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus. J Virol. 2012;86(7):3995–4008. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
