Integrated Analysis of Whole Genome and Epigenome Data Using Machine Learning Technology: Toward the Establishment of Precision Oncology
- PMID: 34055633
- PMCID: PMC8149908
- DOI: 10.3389/fonc.2021.666937
Integrated Analysis of Whole Genome and Epigenome Data Using Machine Learning Technology: Toward the Establishment of Precision Oncology
Abstract
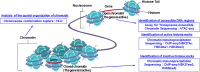
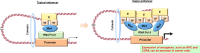
With the completion of the International Human Genome Project, we have entered what is known as the post-genome era, and efforts to apply genomic information to medicine have become more active. In particular, with the announcement of the Precision Medicine Initiative by U.S. President Barack Obama in his State of the Union address at the beginning of 2015, "precision medicine," which aims to divide patients and potential patients into subgroups with respect to disease susceptibility, has become the focus of worldwide attention. The field of oncology is also actively adopting the precision oncology approach, which is based on molecular profiling, such as genomic information, to select the appropriate treatment. However, the current precision oncology is dominated by a method called targeted-gene panel (TGP), which uses next-generation sequencing (NGS) to analyze a limited number of specific cancer-related genes and suggest optimal treatments, but this method causes the problem that the number of patients who benefit from it is limited. In order to steadily develop precision oncology, it is necessary to integrate and analyze more detailed omics data, such as whole genome data and epigenome data. On the other hand, with the advancement of analysis technologies such as NGS, the amount of data obtained by omics analysis has become enormous, and artificial intelligence (AI) technologies, mainly machine learning (ML) technologies, are being actively used to make more efficient and accurate predictions. In this review, we will focus on whole genome sequencing (WGS) analysis and epigenome analysis, introduce the latest results of omics analysis using ML technologies for the development of precision oncology, and discuss the future prospects.
Keywords: artificial intelligence; biomarker discovery; cancer diagnosis and treatment; epigenome analysis; machine learning; precision oncology; whole genome analysis.
Copyright © 2021 Asada, Kaneko, Takasawa, Machino, Takahashi, Shinkai, Shimoyama, Komatsu and Hamamoto.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

