Toward QbD Process Understanding on DNA Vaccine Purification Using Design of Experiment
- PMID: 34055759
- PMCID: PMC8153680
- DOI: 10.3389/fbioe.2021.657201
Toward QbD Process Understanding on DNA Vaccine Purification Using Design of Experiment
Abstract
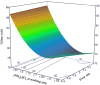
DNA vaccines, the third generation of vaccines, are a promising therapeutic option for many diseases as they offer the customization of their ability on protection and treatment with high stability. The production of DNA vaccines is considered rapid and less complicated compared to others such as mRNA vaccines, viral vaccines, or subunit protein vaccines. However, the main issue for DNA vaccines is how to produce the active DNA, a supercoiled isoform, to comply with the regulations. Our work therefore focuses on gaining a process understanding of the purification step which processes parameters that have impacts on the critical quality attribute (CQA), supercoiled DNA and performance attribute (PA), and step yield. Herein, pVax1/lacZ was used as a model. The process parameters of interest were sample application flow rates and salt concentration at washing step and at elution step in the hydrophobic interaction chromatography (HIC). Using a Design of Experiment (DoE) with central composite face centered (CCF) approach, 14 experiments plus four additional runs at the center points were created. The response data was used to establish regression predictive models and simulation was conducted in 10,000 runs to provide tolerance intervals of these CQA and PA. The approach of this process understanding can be applied for Quality by Design (QbD) on other DNA vaccines and on a larger production scale as well.
Keywords: DNA vaccine purification; Design of Experiment; QbD; process understanding; tolerance study.
Copyright © 2021 Hocharoen, Noppiboon and Kitsubun.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Process Characterization by Definitive Screening Design Approach on DNA Vaccine Production.Front Bioeng Biotechnol. 2020 Oct 15;8:574809. doi: 10.3389/fbioe.2020.574809. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33178673 Free PMC article.
-
Modeling of hydrophobic interaction chromatography for the separation of antibody-drug conjugates and its application towards quality by design.J Biotechnol. 2020 Jun 20;317:48-58. doi: 10.1016/j.jbiotec.2020.04.018. Epub 2020 May 1. J Biotechnol. 2020. PMID: 32361022
-
Design of experiments applications in bioprocessing: Chromatography process development using split design of experiments.Biotechnol Prog. 2019 Jan;35(1):e2730. doi: 10.1002/btpr.2730. Epub 2018 Nov 4. Biotechnol Prog. 2019. PMID: 30315679
-
Application of quality by design in the current drug development.Asian J Pharm Sci. 2017 Jan;12(1):1-8. doi: 10.1016/j.ajps.2016.07.006. Epub 2016 Aug 4. Asian J Pharm Sci. 2017. PMID: 32104308 Free PMC article. Review.
-
Vaccine process technology.Biotechnol Bioeng. 2012 Jun;109(6):1443-60. doi: 10.1002/bit.24493. Epub 2012 Mar 30. Biotechnol Bioeng. 2012. PMID: 22407777 Review.
Cited by
-
Intranasal Ion-Triggered In Situ Delivery System of Virus-like Particles: Development Using the Quality by Design Approach.Polymers (Basel). 2024 Mar 2;16(5):685. doi: 10.3390/polym16050685. Polymers (Basel). 2024. PMID: 38475368 Free PMC article.
-
COMPARING vaccine manufacturing technologies recombinant DNA vs in vitro transcribed (IVT) mRNA.Sci Rep. 2024 Sep 18;14(1):21742. doi: 10.1038/s41598-024-67797-x. Sci Rep. 2024. PMID: 39289418 Free PMC article.
-
Maximization of the Minicircle DNA Vaccine Production Expressing SARS-CoV-2 RBD.Biomedicines. 2022 Apr 25;10(5):990. doi: 10.3390/biomedicines10050990. Biomedicines. 2022. PMID: 35625727 Free PMC article.
-
Design of Experiments to Achieve an Efficient Chitosan-Based DNA Vaccine Delivery System.Pharmaceutics. 2021 Aug 31;13(9):1369. doi: 10.3390/pharmaceutics13091369. Pharmaceutics. 2021. PMID: 34575445 Free PMC article.
References
-
- Azevedo G. M., Valente J. F. A., Sousa A., Pedro A. Q., Pereira P., Sousa F., et al. (2019). Effect of chromatographic conditions on supercoiled plasmid DNA stability and bioactivity. Appl. Sci. 9:5170. 10.3390/app9235170 - DOI
-
- Box G. E. P., Wilson K. B. (1951). On the experimental attainment of optimum conditions. J. R. Stat. Soc. Ser. B 13 1–45. 10.1111/j.2517-6161.1951.tb00067.x - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

