Insights into Aldehyde Dehydrogenase Enzymes: A Structural Perspective
- PMID: 34055881
- PMCID: PMC8160307
- DOI: 10.3389/fmolb.2021.659550
Insights into Aldehyde Dehydrogenase Enzymes: A Structural Perspective
Abstract
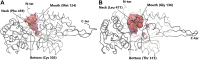
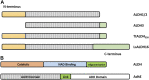
Aldehyde dehydrogenases engage in many cellular functions, however their dysfunction resulting in accumulation of their substrates can be cytotoxic. ALDHs are responsible for the NAD(P)-dependent oxidation of aldehydes to carboxylic acids, participating in detoxification, biosynthesis, antioxidant and regulatory functions. Severe diseases, including alcohol intolerance, cancer, cardiovascular and neurological diseases, were linked to dysfunctional ALDH enzymes, relating back to key enzyme structure. An in-depth understanding of the ALDH structure-function relationship and mechanism of action is key to the understanding of associated diseases. Principal structural features 1) cofactor binding domain, 2) active site and 3) oligomerization mechanism proved critical in maintaining ALDH normal activity. Emerging research based on the combination of structural, functional and biophysical studies of bacterial and eukaryotic ALDHs contributed to the appreciation of diversity within the superfamily. Herewith, we discuss these studies and provide our interpretation for a global understanding of ALDH structure and its purpose-including correct function and role in disease. Our analysis provides a synopsis of a common structure-function relationship to bridge the gap between the highly studied human ALDHs and lesser so prokaryotic models.
Keywords: C-terminal extensions; NAD(P) cofactor; aldehyde dehydrogenase; enzyme dysfunction; mutations; oligomerization; spirosomes; structure-function.
Copyright © 2021 Shortall, Djeghader, Magner and Soulimane.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
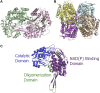

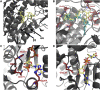
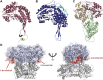

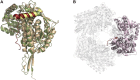

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

