Strategies for Targeting Retroviral Integration for Safer Gene Therapy: Advances and Challenges
- PMID: 34055882
- PMCID: PMC8149907
- DOI: 10.3389/fmolb.2021.662331
Strategies for Targeting Retroviral Integration for Safer Gene Therapy: Advances and Challenges
Abstract
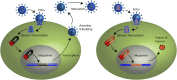
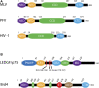
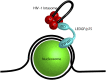
Retroviruses are obligate intracellular parasites that must integrate a copy of the viral genome into the host DNA. The integration reaction is performed by the viral enzyme integrase in complex with the two ends of the viral cDNA genome and yields an integrated provirus. Retroviral vector particles are attractive gene therapy delivery tools due to their stable integration. However, some retroviral integration events may dysregulate host oncogenes leading to cancer in gene therapy patients. Multiple strategies to target retroviral integration, particularly to genetic safe harbors, have been tested with limited success. Attempts to target integration may be limited by the multimerization of integrase or the presence of host co-factors for integration. Several retroviral integration complexes have evolved a mechanism of tethering to chromatin via a host protein. Integration host co-factors bind chromatin, anchoring the complex and allowing integration. The tethering factor allows for both close proximity to the target DNA and specificity of targeting. Each retrovirus appears to have distinct preferences for DNA sequence and chromatin features at the integration site. Tethering factors determine the preference for chromatin features, but do not affect the subtle sequence preference at the integration site. The sequence preference is likely intrinsic to the integrase protein. New developments may uncouple the requirement for a tethering factor and increase the ability to redirect retroviral integration.
Keywords: HIV-1; LEDGF/p75; MLV; gene therapy; retrovirus; targeted integration.
Copyright © 2021 Yoder, Rabe, Fishel and Larue.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

