A Class of Valuable (Pro-)Activity-Based Protein Profiling Probes: Application to the Redox-Active Antiplasmodial Agent, Plasmodione
- PMID: 34056636
- PMCID: PMC8154199
- DOI: 10.1021/jacsau.1c00025
A Class of Valuable (Pro-)Activity-Based Protein Profiling Probes: Application to the Redox-Active Antiplasmodial Agent, Plasmodione
Abstract
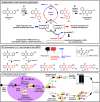
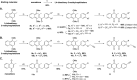
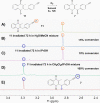
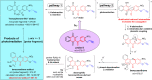
Plasmodione (PD) is a potent antimalarial redox-active drug acting at low nM range concentrations on different malaria parasite stages. In this study, in order to determine the precise PD protein interactome in parasites, we developed a class of (pro-)activity-based protein profiling probes (ABPP) as precursors of photoreactive benzophenone-like probes based on the skeleton of PD metabolites (PDO) generated in a cascade of redox reactions. Under UV-photoirradiation, we clearly demonstrate that benzylic oxidation of 3-benzylmenadione 11 produces the 3-benzoylmenadione probe 7, allowing investigation of the proof-of-concept of the ABPP strategy with 3-benzoylmenadiones 7-10. The synthesized 3-benzoylmenadiones, probe 7 with an alkyne group or probe 9 with -NO2 in para position of the benzoyl chain, were found to be the most efficient photoreactive and clickable probes. In the presence of various H-donor partners, the UV-irradiation of the photoreactive ABPP probes generates different adducts, the expected "benzophenone-like" adducts (pathway 1) in addition to "benzoxanthone" adducts (via two other pathways, 2 and 3). Using both human and Plasmodium falciparum glutathione reductases, three protein ligand binding sites were identified following photolabeling with probes 7 or 9. The photoreduction of 3-benzoylmenadiones (PDO and probe 9) promoting the formation of both the corresponding benzoxanthone and the derived enone could be replaced by the glutathione reductase-catalyzed reduction step. In particular, the electrophilic character of the benzoxanthone was evidenced by its ability to alkylate heme, as a relevant event supporting the antimalarial mode of action of PD. This work provides a proof-of-principle that (pro-)ABPP probes can generate benzophenone-like metabolites enabling optimized activity-based protein profiling conditions that will be instrumental to analyze the interactome of early lead antiplasmodial 3-benzylmenadiones displaying an original and innovative mode of action.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures








References
-
- Yoo E.; Schulze C. J.; Stokes B. H.; Onguka O.; Yeo T.; Mok S.; Gnädig N. F.; Zhou Y.; Kurita K.; Foe I. T.; Terrell S. M.; Boucher M. J.; Cieplak P.; Kumpornsin K.; Lee M. C. S.; Linington R. G.; Long J. Z.; Uhlemann A. C.; Weerapana E.; Fidock D. A.; Bogyo M. The Antimalarial Natural Product Salinipostin A Identifies Essential α/β Serine Hydrolases Involved in Lipid Metabolism in P. Falciparum Parasites. Cell Chem. Biol. 2020, 27, 143–157. 10.1016/j.chembiol.2020.01.001. - DOI - PMC - PubMed
-
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem., Int. Ed. 2002, 41, 2596–2599. 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
