Cytogenetic events in the endosperm of amphiploid Avena magna × A. longiglumis
- PMID: 34057611
- PMCID: PMC8364899
- DOI: 10.1007/s10265-021-01314-3
Cytogenetic events in the endosperm of amphiploid Avena magna × A. longiglumis
Abstract
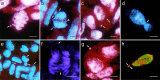
This study analysed cytogenetic events occurring in the syncytial endosperm of the Avena magna H. C. Murphy & Terrell × Avena longiglumis Durieu amphiploid, which is a product of two wild species having different genomes. Selection through the elimination of chromosomes and their fragments, including those translocated, decreased the level of ploidy in the endosperm below the expected 3n, leading to the modal number close to 2n. During intergenomic translocations, fragments of the heterochromatin-rich C-genome were transferred to the D and Al genomes. Terminal and non-reciprocal exchanges dominated, whereas other types of translocations, including microexchanges, were less common. Using two probes and by counterstaining with DAPI, the A. longiglumis and the rare exchanges between the D and Al genomes were detected by GISH. The large discontinuity in the probe labelling in the C chromosomes demonstrated inequality in the distribution of repetitive sequences along the chromosome and probable intragenomic rearrangements. In the nucleus, the spatial arrangement of genomes was non-random and showed a sectorial-concentric pattern, which can vary during the cell cycle, especially in the less stable tissue like the hybrid endosperm.
Keywords: Amphiploid; Avena; Chromosome rearrangements; Genome domains; Nuclear disorders.
© 2021. The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
The repetitive DNA landscape in Avena (Poaceae): chromosome and genome evolution defined by major repeat classes in whole-genome sequence reads.BMC Plant Biol. 2019 May 30;19(1):226. doi: 10.1186/s12870-019-1769-z. BMC Plant Biol. 2019. PMID: 31146681 Free PMC article.
-
Chromosomal distribution patterns of the (AC)10 microsatellite and other repetitive sequences, and their use in chromosome rearrangement analysis of species of the genus Avena.Genome. 2017 Mar;60(3):216-227. doi: 10.1139/gen-2016-0146. Epub 2016 Nov 4. Genome. 2017. PMID: 28156137
-
The mitochondrial genome of the diploid oat Avena longiglumis.BMC Plant Biol. 2023 Apr 26;23(1):218. doi: 10.1186/s12870-023-04217-8. BMC Plant Biol. 2023. PMID: 37098475 Free PMC article.
-
Chromosome-level genome assembly of the diploid oat species Avena longiglumis.Sci Data. 2024 Apr 22;11(1):412. doi: 10.1038/s41597-024-03248-6. Sci Data. 2024. PMID: 38649380 Free PMC article.
-
A new chromosome nomenclature system for oat (Avena sativa L. and A. byzantina C. Koch) based on FISH analysis of monosomic lines.Theor Appl Genet. 2010 Nov;121(8):1541-52. doi: 10.1007/s00122-010-1409-3. Epub 2010 Jul 24. Theor Appl Genet. 2010. PMID: 20658121
Cited by
-
Complex polyploid and hybrid species in an apomictic and sexual tropical forage grass group: genomic composition and evolution in Urochloa (Brachiaria) species.Ann Bot. 2023 Feb 7;131(1):87-108. doi: 10.1093/aob/mcab147. Ann Bot. 2023. PMID: 34874999 Free PMC article.
-
Oat chromosome and genome evolution defined by widespread terminal intergenomic translocations in polyploids.Front Plant Sci. 2022 Nov 22;13:1026364. doi: 10.3389/fpls.2022.1026364. eCollection 2022. Front Plant Sci. 2022. PMID: 36483968 Free PMC article.
-
Oat species and interspecific amphiploids show predominance of diploid nuclei in the syncytial endosperm.J Appl Genet. 2024 Feb;65(1):1-11. doi: 10.1007/s13353-023-00798-0. Epub 2023 Nov 7. J Appl Genet. 2024. PMID: 37934380 Free PMC article.
References
-
- An LH, You RL. Studies on nuclear degeneration during programmed cell death of synergid and antipodal cells in Triticum aestivum. Sex Plant Reprod. 2004;17:195–201. doi: 10.1007/s00497-004-0220-1. - DOI
-
- Baptista-Giacomelli FR, Pagliarini MS, de Almeida JL. Elimination of micronuclei from microspores in a Brazilian oat (Avena sativa L.) variety. Genet Mol Biol. 2000 doi: 10.1590/S1415-47572000000300029. - DOI
-
- Bennett MD. Nuclear architecture and its manipulation. In: Gustafson JP, editor. Gene manipulation in plant improvement, Stadler genetics symposia series. Boston: Springer; 1984. pp. 469–502.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

