Inflammasome regulation in driving COVID-19 severity in humans and immune tolerance in bats
- PMID: 34057760
- PMCID: PMC8242921
- DOI: 10.1002/JLB.4COVHR0221-093RR
Inflammasome regulation in driving COVID-19 severity in humans and immune tolerance in bats
Abstract
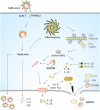
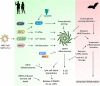
Coronaviruses (CoVs) are RNA viruses that cause human respiratory infections. Zoonotic transmission of the SARS-CoV-2 virus caused the recent COVID-19 pandemic, which led to over 2 million deaths worldwide. Elevated inflammatory responses and cytotoxicity in the lungs are associated with COVID-19 severity in SARS-CoV-2-infected individuals. Bats, which host pathogenic CoVs, operate dampened inflammatory responses and show tolerance to these viruses with mild clinical symptoms. Delineating the mechanisms governing these host-specific inflammatory responses is essential to understand host-virus interactions determining the outcome of pathogenic CoV infections. Here, we describe the essential role of inflammasome activation in determining COVID-19 severity in humans and innate immune tolerance in bats that host several pathogenic CoVs. We further discuss mechanisms leading to inflammasome activation in human SARS-CoV-2 infection and how bats are molecularly adapted to suppress these inflammasome responses. We also report an analysis of functionally important residues of inflammasome components that provide new clues of bat strategies to suppress inflammasome signaling and innate immune responses. As spillover of bat viruses may cause the emergence of new human disease outbreaks, the inflammasome regulation in bats and humans likely provides specific strategies to combat the pathogenic CoV infections.
Keywords: Bat immunity; COVID-19; Coronaviruses; Immune tolerance; Inflammasome; NLRP3; SARS-CoV-2.
©2021 Society for Leukocyte Biology.
Figures


References
-
- Lan J, Ge J, Yu J, et al. Structure of the SARS‐CoV‐2 spike receptor‐binding domain bound to the ACE2 receptor. Nature. 2020;581:215‐220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

