Immune characterization and profiles of SARS-CoV-2 infected patients reveals potential host therapeutic targets and SARS-CoV-2 oncogenesis mechanism
- PMID: 34058265
- PMCID: PMC8163696
- DOI: 10.1016/j.virusres.2021.198464
Immune characterization and profiles of SARS-CoV-2 infected patients reveals potential host therapeutic targets and SARS-CoV-2 oncogenesis mechanism
Abstract
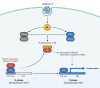
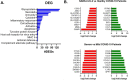
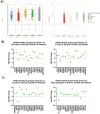
The spread of SARS-CoV-2 and the increasing mortality rates of COVID-19 create an urgent need for treatments, which are currently lacking. Although vaccines have been approved by the FDA for emergency use in the U.S., patients will continue to require pharmacologic intervention to reduce morbidity and mortality as vaccine availability remains limited. The rise of new variants makes the development of therapeutic strategies even more crucial to combat the current pandemic and future outbreaks. Evidence from several studies suggests the host immune response to SARS-CoV-2 infection plays a critical role in disease pathogenesis. Consequently, host immune factors are becoming more recognized as potential biomarkers and therapeutic targets for COVID-19. To develop therapeutic strategies to combat current and future coronavirus outbreaks, understanding how the coronavirus hijacks the host immune system during and after the infection is crucial. In this study, we investigated immunological patterns or characteristics of the host immune response to SARS-CoV-2 infection that may contribute to the disease severity of COVID-19 patients. We analyzed large bulk RNASeq and single cell RNAseq data from COVID-19 patient samples to immunoprofile differentially expressed gene sets and analyzed pathways to identify human host protein targets. We observed an immunological profile of severe COVID-19 patients characterized by upregulated cytokines, interferon-induced proteins, and pronounced T cell lymphopenia, supporting findings by previous studies. We identified a number of host immune targets including PERK, PKR, TNF, NF-kB, and other key genes that modulate the significant pathways and genes identified in COVID-19 patients. Finally, we identified genes modulated by COVID-19 infection that are implicated in oncogenesis, including E2F transcription factors and RB1, suggesting a mechanism by which SARS-CoV-2 infection may contribute to oncogenesis. Further clinical investigation of these targets may lead to bonafide therapeutic strategies to treat the current COVID-19 pandemic and protect against future outbreaks and viral escape variants.
Keywords: Bulk RNASeq analysis; Cancer; Gene expression analysis; Immune characterization of COVID19 patients; Immune profiling of COVID19 patients; SARS-COV-2; Single cell scRNAseq analysis.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

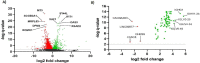




Update of
-
Immune characterization and profiles of SARS-CoV-2 infected patients reveals potential host therapeutic targets and SARS-CoV-2 oncogenesis mechanism.bioRxiv [Preprint]. 2021 Mar 22:2021.02.17.431721. doi: 10.1101/2021.02.17.431721. bioRxiv. 2021. Update in: Virus Res. 2021 Aug;301:198464. doi: 10.1016/j.virusres.2021.198464. PMID: 33619493 Free PMC article. Updated. Preprint.
Similar articles
-
Immune characterization and profiles of SARS-CoV-2 infected patients reveals potential host therapeutic targets and SARS-CoV-2 oncogenesis mechanism.bioRxiv [Preprint]. 2021 Mar 22:2021.02.17.431721. doi: 10.1101/2021.02.17.431721. bioRxiv. 2021. Update in: Virus Res. 2021 Aug;301:198464. doi: 10.1016/j.virusres.2021.198464. PMID: 33619493 Free PMC article. Updated. Preprint.
-
Comparative analysis of dynamic transcriptomes reveals specific COVID-19 features and pathogenesis of immunocompromised populations.mSystems. 2024 Jun 18;9(6):e0138523. doi: 10.1128/msystems.01385-23. Epub 2024 May 16. mSystems. 2024. PMID: 38752789 Free PMC article.
-
Transcriptome of nasopharyngeal samples from COVID-19 patients and a comparative analysis with other SARS-CoV-2 infection models reveal disparate host responses against SARS-CoV-2.J Transl Med. 2021 Jan 7;19(1):32. doi: 10.1186/s12967-020-02695-0. J Transl Med. 2021. PMID: 33413422 Free PMC article.
-
Insights into COVID-19 Vaccine Development Based on Immunogenic Structural Proteins of SARS-CoV-2, Host Immune Responses, and Herd Immunity.Cells. 2021 Oct 29;10(11):2949. doi: 10.3390/cells10112949. Cells. 2021. PMID: 34831172 Free PMC article. Review.
-
Potential Therapeutic Targets and Vaccine Development for SARS-CoV-2/COVID-19 Pandemic Management: A Review on the Recent Update.Front Immunol. 2021 Jun 30;12:658519. doi: 10.3389/fimmu.2021.658519. eCollection 2021. Front Immunol. 2021. PMID: 34276652 Free PMC article. Review.
Cited by
-
Novel risk loci for COVID-19 hospitalization among admixed American populations.Elife. 2024 Oct 3;13:RP93666. doi: 10.7554/eLife.93666. Elife. 2024. PMID: 39361370 Free PMC article.
-
First encounter with SARS-CoV-2: immune portraits of COVID susceptibility.Nature. 2024 Jul;631(8019):33-35. doi: 10.1038/d41586-024-01644-x. Nature. 2024. PMID: 38898251 No abstract available.
-
SARS-CoV-2 Infection: A Possible Risk Factor for Incidence and Recurrence of Cancers.Int J Hematol Oncol Stem Cell Res. 2022 Apr 1;16(2):117-127. doi: 10.18502/ijhoscr.v16i2.9205. Int J Hematol Oncol Stem Cell Res. 2022. PMID: 36304732 Free PMC article. Review.
-
Tryptophan-Kynurenine Pathway in COVID-19-Dependent Musculoskeletal Pathology: A Minireview.Mediators Inflamm. 2021 Oct 5;2021:2911578. doi: 10.1155/2021/2911578. eCollection 2021. Mediators Inflamm. 2021. PMID: 34621138 Free PMC article. Review.
-
Characterizing the key genes of COVID-19 that regulate tumor immune microenvironment and prognosis in hepatocellular carcinoma.Funct Integr Genomics. 2023 Aug 4;23(3):262. doi: 10.1007/s10142-023-01184-z. Funct Integr Genomics. 2023. PMID: 37540264
References
-
- Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., Soboleva A. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. Database issue. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

