Hypoxia-induced lncHILAR promotes renal cancer metastasis via ceRNA for the miR-613/206/ 1-1-3p/Jagged-1/Notch/CXCR4 signaling pathway
- PMID: 34058384
- PMCID: PMC8531137
- DOI: 10.1016/j.ymthe.2021.05.020
Hypoxia-induced lncHILAR promotes renal cancer metastasis via ceRNA for the miR-613/206/ 1-1-3p/Jagged-1/Notch/CXCR4 signaling pathway
Retraction in
-
Retraction Notice to: Hypoxia-induced lncHILAR promotes renal cancer metastasis via ceRNA for the miR-613/206/ 1-1-3p/Jagged-1/Notch/CXCR4 signaling pathway.Mol Ther. 2025 Mar 5;33(3):1303. doi: 10.1016/j.ymthe.2025.01.044. Epub 2025 Jan 31. Mol Ther. 2025. PMID: 39892388 Free PMC article. No abstract available.
Abstract
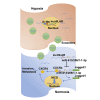
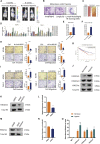
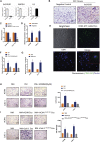
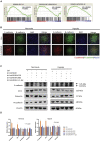
Hypoxia has been identified as a common contributor to tumor progression, including invasion and metastasis. However, the underlying mechanisms of enhanced invasion and metastasis under hypoxia remain unclear. A hypoxic microenvironment promotes invasion and metastasis of renal cell carcinoma (RCC) by upregulating expression of LOC100506178, which we named hypoxia-induced long non-coding RNA (lncRNA) associated with RCC (lncHILAR). Knockdown of lncHILAR inhibited cell invasion and migration, whereas overexpression of lncHILAR, conversely, facilitated cell invasion and migration of RCC cells. Notably, hypoxic RCC cells secreted exosomes packaged with lncHILAR, which were taken up by normoxic RCC cells and then drove normoxic cell invasion. Mechanistically, lncHILAR elevated RCC invasion and metastasis by acting as a competing endogenous RNA (ceRNA) for miR-613/206/1-1-3p, which led to the upregulation of Jagged-1 and the C-X-C motif chemokine receptor 4 (CXCR4). Activation of the Jagged-1/Notch/CXCR4 axis induced RCC metastasis. lncHILAR promotes RCC cell invasion and metastasis via ceRNA for the miR-613/206/1-1-3p/Jagged-1/Notch/CXCR4 axis. The novel lncHILAR may thus serve as a potential biomarker and therapeutic target in RCC.
Keywords: RCC; exosomes; hypoxia; lncRNA; metastasis.
Copyright © 2021 The American Society of Gene and Cell Therapy. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



Similar articles
-
Long noncoding RNA lung cancer associated transcript 1 promotes proliferation and invasion of clear cell renal cell carcinoma cells by negatively regulating miR-495-3p.J Cell Biochem. 2018 Sep;119(9):7599-7609. doi: 10.1002/jcb.27099. Epub 2018 Jun 22. J Cell Biochem. 2018. PMID: 29932248
-
Depletion of lncRNA MALAT1 inhibited sunitinib resistance through regulating miR-362-3p-mediated G3BP1 in renal cell carcinoma.Cell Cycle. 2020 Aug;19(16):2054-2062. doi: 10.1080/15384101.2020.1792667. Epub 2020 Jul 14. Cell Cycle. 2020. PMID: 32663095 Free PMC article.
-
LncRNA FGD5-AS1/miR-5590-3p axis facilitates the proliferation and metastasis of renal cell carcinoma through ERK/AKT signalling.Eur Rev Med Pharmacol Sci. 2020 Sep;24(17):8756-8766. doi: 10.26355/eurrev_202009_22814. Eur Rev Med Pharmacol Sci. 2020. PMID: 32964964
-
ciRS-7 is a prognostic biomarker and potential gene therapy target for renal cell carcinoma.Mol Cancer. 2021 Nov 5;20(1):142. doi: 10.1186/s12943-021-01443-2. Mol Cancer. 2021. PMID: 34740354 Free PMC article.
-
MYOSLID: A Critical Modulator of Cancer Hallmarks.Genes (Basel). 2025 Mar 14;16(3):341. doi: 10.3390/genes16030341. Genes (Basel). 2025. PMID: 40149492 Free PMC article. Review.
Cited by
-
Loss of MIR503HG facilitates papillary renal cell carcinoma associated lymphatic metastasis by triggering NOTCH1/VEGFC signaling.Int J Biol Sci. 2023 Jun 19;19(10):3266-3284. doi: 10.7150/ijbs.83302. eCollection 2023. Int J Biol Sci. 2023. PMID: 37416763 Free PMC article.
-
Extracellular putrescine can augment the epithelial-mesenchymal transition of gastric cancer cells by promoting MAL2 expression by elevating H3K27ac in its promoter region.Am J Cancer Res. 2024 Jun 15;14(6):2805-2822. doi: 10.62347/BEUV4081. eCollection 2024. Am J Cancer Res. 2024. PMID: 39005660 Free PMC article.
-
LncRNA profiles from Notch signaling: Implications for clinical management and tumor microenvironment of colorectal cancer.Front Immunol. 2022 Jul 25;13:953405. doi: 10.3389/fimmu.2022.953405. eCollection 2022. Front Immunol. 2022. PMID: 35958606 Free PMC article.
-
Molecular mechanisms of renal cell carcinoma metastasis and potential targets for therapy.Front Cell Dev Biol. 2025 Jan 20;13:1521151. doi: 10.3389/fcell.2025.1521151. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 39901876 Free PMC article. Review.
-
Identification of Hypoxia-Related Prognostic Signature and Competing Endogenous RNA Regulatory Axes in Hepatocellular Carcinoma.Int J Mol Sci. 2022 Nov 5;23(21):13590. doi: 10.3390/ijms232113590. Int J Mol Sci. 2022. PMID: 36362375 Free PMC article.
References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. - PubMed
-
- Khanna A., Crane A., Yerram N., Sun D., Ericson K., Lundy S.D., Abouassaly R. Contemporary management of advanced renal cell carcinoma. Clin. Adv. Hematol. Oncol. 2018;16:438–446. - PubMed
-
- Schito L., Semenza G.L. Hypoxia-inducible factors: master regulators of cancer progression. Trends Cancer. 2016;2:758–770. - PubMed
-
- Deng S.J., Chen H.Y., Ye Z., Deng S.C., Zhu S., Zeng Z., He C., Liu M.L., Huang K., Zhong J.X. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811–5828. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

