Correlating ion channel structure and function
- PMID: 34059287
- PMCID: PMC9288845
- DOI: 10.1016/bs.mie.2021.02.016
Correlating ion channel structure and function
Abstract
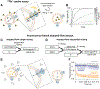
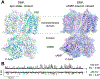
Recent developments in cryogenic electron microscopy (cryo-EM) led to an exponential increase in high-resolution structures of membrane proteins, and in particular ion channels. However, structures alone can only provide limited information about the workings of these proteins. In order to understand ion channel function and regulation in molecular detail, the obtained structural data need to be correlated to functional states of the same protein. Here, we describe several techniques that can be employed to study ion channel structure and function in vitro and under defined, similar conditions. Lipid nanodiscs provide a native-like environment for membrane proteins and have become a valuable tool in membrane protein structural biology and biophysics. Combined with liposome-based flux assays for the kinetic analysis of ion channel activity as well as electrophysiological recordings, researchers now have access to an array of experimental techniques allowing for detailed structure-function correlations using purified components. Two examples are presented where we put emphasis on the lipid environment and time-resolved techniques together with mutations and protein engineering to interpret structural data obtained from single particle cryo-EM on cyclic nucleotide-gated or Ca2+-gated K+ channels. Furthermore, we provide short protocols for all the assays used in our work so that others can adapt these techniques to their experimental needs. Comprehensive structure-function correlations are essential in order to pharmacologically target channelopathies.
Keywords: Cryo-electron microscopy; Ion channel; Lipid bilayers; MthK; Nanodisc; Radioactive uptake assay; Single-channel recording; SthK; Stopped-flow fluorescence assay.
© 2021 Elsevier Inc. All rights reserved.
Figures

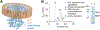
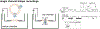
Similar articles
-
Lipid Nanodiscs as a Tool for High-Resolution Structure Determination of Membrane Proteins by Single-Particle Cryo-EM.Methods Enzymol. 2017;594:1-30. doi: 10.1016/bs.mie.2017.05.007. Epub 2017 Jul 19. Methods Enzymol. 2017. PMID: 28779836
-
Ligand discrimination and gating in cyclic nucleotide-gated ion channels from apo and partial agonist-bound cryo-EM structures.Elife. 2018 Jul 20;7:e39775. doi: 10.7554/eLife.39775. Elife. 2018. PMID: 30028291 Free PMC article.
-
Lipid nanodisc scaffold and size alter the structure of a pentameric ligand-gated ion channel.Nat Commun. 2024 Jan 2;15(1):25. doi: 10.1038/s41467-023-44366-w. Nat Commun. 2024. PMID: 38167383 Free PMC article.
-
Determining the Crystal Structure of TRPV6.In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 14. In: Kozak JA, Putney JW Jr, editors. Calcium Entry Channels in Non-Excitable Cells. Boca Raton (FL): CRC Press/Taylor & Francis; 2018. Chapter 14. PMID: 30299652 Free Books & Documents. Review.
-
To Be or Not to Be an Ion Channel: Cryo-EM Structures Have a Say.Cells. 2023 Jul 17;12(14):1870. doi: 10.3390/cells12141870. Cells. 2023. PMID: 37508534 Free PMC article. Review.
Cited by
-
Domain Coupling in Allosteric Regulation of SthK Measured Using Time-Resolved Transition Metal Ion FRET.bioRxiv [Preprint]. 2025 May 20:2025.03.31.646362. doi: 10.1101/2025.03.31.646362. bioRxiv. 2025. Update in: Elife. 2025 Aug 12;14:RP106892. doi: 10.7554/eLife.106892. PMID: 40236086 Free PMC article. Updated. Preprint.
-
Rapid Multi-Well Evaluation of Assorted Materials for Hydrogel-Assisted Giant Unilamellar Vesicle Production: Empowering Bottom-Up Synthetic Biology.Gels. 2025 Jan 2;11(1):29. doi: 10.3390/gels11010029. Gels. 2025. PMID: 39852000 Free PMC article.
-
Oxidative Stress and Immune Response in Melanoma: Ion Channels as Targets of Therapy.Int J Mol Sci. 2023 Jan 3;24(1):887. doi: 10.3390/ijms24010887. Int J Mol Sci. 2023. PMID: 36614330 Free PMC article. Review.
-
Domain coupling in allosteric regulation of SthK measured using time-resolved transition metal ion FRET.Elife. 2025 Aug 12;14:RP106892. doi: 10.7554/eLife.106892. Elife. 2025. PMID: 40792615 Free PMC article.
-
Native Function of the Bacterial Ion Channel SthK in a Sparsely Tethered Lipid Bilayer Membrane Architecture.J Phys Chem B. 2023 Apr 27;127(16):3641-3650. doi: 10.1021/acs.jpcb.2c07252. Epub 2023 Apr 18. J Phys Chem B. 2023. PMID: 37072125 Free PMC article.
References
-
- Banerjee S, Huber T, and Sakmar TP. 2008. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J Mol Biol. 377:1067–1081. - PubMed
-
- Bayburt TH, Grinkova YV, and Sligar SG. 2002. Self-Assembly of Discoidal Phospholipid Bilayer Nanoparticles with Membrane Scaffold Proteins. Nano Letters. 2:853–856.
-
- Bezanilla F 2008. Ion channels: from conductance to structure. Neuron. 60:456–468. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

