P300/HDAC1 regulates the acetylation/deacetylation and autophagic activities of LC3/Atg8-PE ubiquitin-like system
- PMID: 34059632
- PMCID: PMC8166822
- DOI: 10.1038/s41420-021-00513-0
P300/HDAC1 regulates the acetylation/deacetylation and autophagic activities of LC3/Atg8-PE ubiquitin-like system
Abstract
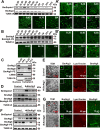
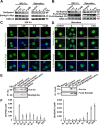
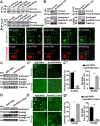
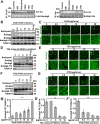
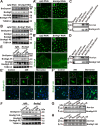
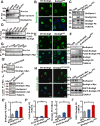
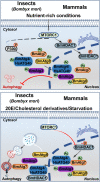
Protein acetylation plays potential roles in regulating autophagy occurrence. However, it varies greatly between yeast and mammals, and has not been thoroughly investigated in other organisms. Here, we reported that the components of BmAtg8-PE ubiquitin-like system (BmAtg3, BmAtg4, BmAtg7, and BmAtg8) in Bombyx mori were localized in the nucleus under nutrient-rich conditions, whereas they were exported to the cytoplasm upon autophagy induction. RNAi of BmP300 and inhibition of BmP300 activity resulted in nucleo-cytoplasmic translocation of BmAtg3 and BmAtg8, as well as premature induction of autophagy in the absence of stimulus. Conversely, RNAi of BmHDAC1 and inhibition of class I/II HADCs activities led to the nuclear accumulation of BmAtg3 and BmAtg8. In addition, acetylation sites in Atg proteins of BmAtg8-PE ubiquitin-like system were identified by mass spectrometry, and acetylation-site mutations caused nucleo-cytoplasmic translocation of BmAtg3, BmAtg4, and BmAtg8 along with autophagy promotion. Similarly, the subcellular localization of human ATG4b is determined by acetylation modification. In general, BmP300-mediated acetylation sequesters the components of BmAtg8-PE ubiquitin-like system in the nucleus, thus leading to the autophagy inhibition. Oppositely, BmHDAC1-mediated deacetylation leads to the nucleo-cytoplasmic translocation of the components of BmAtg8-PE ubiquitin-like system and promotes autophagy. This process is evolutionarily conserved between insects and mammals.
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Acetylation of BmAtg8 inhibits starvation-induced autophagy initiation.Mol Cell Biochem. 2019 Jul;457(1-2):73-81. doi: 10.1007/s11010-019-03513-y. Epub 2019 Mar 15. Mol Cell Biochem. 2019. PMID: 30877510
-
Proteasome inhibitor MG132 impairs autophagic flux through compromising formation of autophagosomes in Bombyx cells.Biochem Biophys Res Commun. 2016 Oct 28;479(4):690-696. doi: 10.1016/j.bbrc.2016.09.151. Epub 2016 Sep 28. Biochem Biophys Res Commun. 2016. PMID: 27693582
-
Cloning and functional analysis of autophagy-related gene 7 in Bombyx mori, silkworm.Arch Insect Biochem Physiol. 2021 Aug;107(4):e21827. doi: 10.1002/arch.21827. Epub 2021 Jun 25. Arch Insect Biochem Physiol. 2021. PMID: 34173258
-
Two ubiquitin-like conjugation systems that mediate membrane formation during autophagy.Essays Biochem. 2013;55:39-50. doi: 10.1042/bse0550039. Essays Biochem. 2013. PMID: 24070470 Review.
-
The Atg8 family: multifunctional ubiquitin-like key regulators of autophagy.Essays Biochem. 2013;55:51-64. doi: 10.1042/bse0550051. Essays Biochem. 2013. PMID: 24070471 Review.
Cited by
-
Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.Autophagy. 2024 Sep;20(9):1948-1967. doi: 10.1080/15548627.2024.2353443. Epub 2024 May 27. Autophagy. 2024. PMID: 38726830 Free PMC article.
-
Epigenetic targeting of autophagy for cancer: DNA and RNA methylation.Front Oncol. 2023 Dec 8;13:1290330. doi: 10.3389/fonc.2023.1290330. eCollection 2023. Front Oncol. 2023. PMID: 38148841 Free PMC article. Review.
-
The role of protein acetylation in carcinogenesis and targeted drug discovery.Front Endocrinol (Lausanne). 2022 Sep 12;13:972312. doi: 10.3389/fendo.2022.972312. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36171897 Free PMC article. Review.
-
Cytoskeleton Protein BmACT1 Is Potential for the Autophagic Function and Nuclear Localization of BmAtg4b in Bombyx mori.Cells. 2023 Mar 15;12(6):899. doi: 10.3390/cells12060899. Cells. 2023. PMID: 36980240 Free PMC article.
-
SETD7 promotes LC3B methylation and degradation in ovarian cancer.J Biol Chem. 2025 Feb;301(2):108134. doi: 10.1016/j.jbc.2024.108134. Epub 2024 Dec 25. J Biol Chem. 2025. PMID: 39725038 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

