The SOC1-like gene BoMADS50 is associated with the flowering of Bambusa oldhamii
- PMID: 34059654
- PMCID: PMC8166863
- DOI: 10.1038/s41438-021-00557-4
The SOC1-like gene BoMADS50 is associated with the flowering of Bambusa oldhamii
Abstract
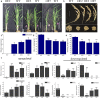
Bamboo is known for its edible shoots and beautiful texture and has considerable economic and ornamental value. Unique among traditional flowering plants, many bamboo plants undergo extensive synchronized flowering followed by large-scale death, seriously affecting the productivity and application of bamboo forests. To date, the molecular mechanism of bamboo flowering characteristics has remained unknown. In this study, a SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1)-like gene, BoMADS50, was identified from Bambusa oldhamii. BoMADS50 was highly expressed in mature leaves and the floral primordium formation period during B. oldhamii flowering and overexpression of BoMADS50 caused early flowering in transgenic rice. Moreover, BoMADS50 could interact with APETALA1/FRUITFULL (AP1/FUL)-like proteins (BoMADS14-1/2, BoMADS15-1/2) in vivo, and the expression of BoMADS50 was significantly promoted by BoMADS14-1, further indicating a synergistic effect between BoMADS50 and BoAP1/FUL-like proteins in regulating B. oldhamii flowering. We also identified four additional transcripts of BoMADS50 (BoMADS50-1/2/3/4) with different nucleotide variations. Although the protein-CDS were polymorphic, they had flowering activation functions similar to those of BoMADS50. Yeast one-hybrid and transient expression assays subsequently showed that both BoMADS50 and BoMADS50-1 bind to the promoter fragment of itself and the SHORT VEGETATIVE PHASE (SVP)-like gene BoSVP, but only BoMADS50-1 can positively induce their transcription. Therefore, nucleotide variations likely endow BoMADS50-1 with strong regulatory activity. Thus, BoMADS50 and BoMADS50-1/2/3/4 are probably important positive flowering regulators in B. oldhamii. Moreover, the functional conservatism and specificity of BoMADS50 and BoMADS50-1 might be related to the synchronized and sporadic flowering characteristics of B. oldhamii.
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Comparative phylogenomic analyses and co-expression gene network reveal insights in flowering time and aborted meiosis in woody bamboo, Bambusa oldhamii 'Xia Zao' ZSX.Front Plant Sci. 2022 Nov 9;13:1023240. doi: 10.3389/fpls.2022.1023240. eCollection 2022. Front Plant Sci. 2022. PMID: 36438131 Free PMC article.
-
Understanding bamboo flowering based on large-scale analysis of expressed sequence tags.Genet Mol Res. 2010 Jun 11;9(2):1085-93. doi: 10.4238/vol9-2gmr804. Genet Mol Res. 2010. PMID: 20568053
-
Overexpression of PvCO1, a bamboo CONSTANS-LIKE gene, delays flowering by reducing expression of the FT gene in transgenic Arabidopsis.BMC Plant Biol. 2018 Oct 12;18(1):232. doi: 10.1186/s12870-018-1469-0. BMC Plant Biol. 2018. PMID: 30314465 Free PMC article.
-
Ectopic expression of the BoTFL1-like gene of Bambusa oldhamii delays blossoming in Arabidopsis thaliana and rescues the tfl1 mutant phenotype.Genet Mol Res. 2015 Aug 10;14(3):9306-17. doi: 10.4238/2015.August.10.11. Genet Mol Res. 2015. PMID: 26345864
-
Regulation and function of SOC1, a flowering pathway integrator.J Exp Bot. 2010 May;61(9):2247-54. doi: 10.1093/jxb/erq098. Epub 2010 Apr 22. J Exp Bot. 2010. PMID: 20413527 Review.
Cited by
-
Metabolic Profiling and Transcriptome Analysis Reveal the Key Role of Flavonoids in Internode Coloration of Phyllostachys violascens cv. Viridisulcata.Front Plant Sci. 2022 Jan 28;12:788895. doi: 10.3389/fpls.2021.788895. eCollection 2021. Front Plant Sci. 2022. PMID: 35154183 Free PMC article.
-
Homo- and Hetero-Dimers of CAD Enzymes Regulate Lignification and Abiotic Stress Response in Moso Bamboo.Int J Mol Sci. 2021 Nov 29;22(23):12917. doi: 10.3390/ijms222312917. Int J Mol Sci. 2021. PMID: 34884720 Free PMC article.
-
Comparative phylogenomic analyses and co-expression gene network reveal insights in flowering time and aborted meiosis in woody bamboo, Bambusa oldhamii 'Xia Zao' ZSX.Front Plant Sci. 2022 Nov 9;13:1023240. doi: 10.3389/fpls.2022.1023240. eCollection 2022. Front Plant Sci. 2022. PMID: 36438131 Free PMC article.
-
Characterization of Phytohormones and Transcriptomic Profiling of the Female and Male Inflorescence Development in Manchurian Walnut (Juglans mandshurica Maxim.).Int J Mol Sci. 2022 May 13;23(10):5433. doi: 10.3390/ijms23105433. Int J Mol Sci. 2022. PMID: 35628244 Free PMC article.
-
Longer Internode with Same Cell Length: LcSOC1-b2 Gene Involved in Height to First Pod but Not Flowering in Lentil (Lens culinaris Medik.).Plants (Basel). 2025 Apr 8;14(8):1157. doi: 10.3390/plants14081157. Plants (Basel). 2025. PMID: 40284045 Free PMC article.
References
-
- Fang, W. et al. Chinese Economic Bamboo (Science Press, 2015).
-
- Franklin DC. Synchrony and asynchrony: observations and hypotheses for the flowering wave in a long-lived semelparous bamboo. J. Biogeogr. 2010;31:773–786. doi: 10.1111/j.1365-2699.2003.01057.x. - DOI
LinkOut - more resources
Full Text Sources
Research Materials

