Mechanically activated ion channel Piezo1 modulates macrophage polarization and stiffness sensing
- PMID: 34059671
- PMCID: PMC8167181
- DOI: 10.1038/s41467-021-23482-5
Mechanically activated ion channel Piezo1 modulates macrophage polarization and stiffness sensing
Abstract
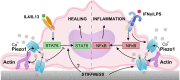
Macrophages perform diverse functions within tissues during immune responses to pathogens and injury, but molecular mechanisms by which physical properties of the tissue regulate macrophage behavior are less well understood. Here, we examine the role of the mechanically activated cation channel Piezo1 in macrophage polarization and sensing of microenvironmental stiffness. We show that macrophages lacking Piezo1 exhibit reduced inflammation and enhanced wound healing responses. Additionally, macrophages expressing the transgenic Ca2+ reporter, Salsa6f, reveal that Ca2+ influx is dependent on Piezo1, modulated by soluble signals, and enhanced on stiff substrates. Furthermore, stiffness-dependent changes in macrophage function, both in vitro and in response to subcutaneous implantation of biomaterials in vivo, require Piezo1. Finally, we show that positive feedback between Piezo1 and actin drives macrophage activation. Together, our studies reveal that Piezo1 is a mechanosensor of stiffness in macrophages, and that its activity modulates polarization responses.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM008620/GM/NIGMS NIH HHS/United States
- R01 AI121945/AI/NIAID NIH HHS/United States
- R21 AR077288/AR/NIAMS NIH HHS/United States
- S10 OD025064/OD/NIH HHS/United States
- R21 AI128519/AI/NIAID NIH HHS/United States
- T32 HL116270/HL/NHLBI NIH HHS/United States
- P30 AR075047/AR/NIAMS NIH HHS/United States
- F30 AI142986/AI/NIAID NIH HHS/United States
- R21 EB027840/EB/NIBIB NIH HHS/United States
- R01 AI151301/AI/NIAID NIH HHS/United States
- R01 NS109810/NS/NINDS NIH HHS/United States
- R01 NS014609/NS/NINDS NIH HHS/United States
- P50 GM076516/GM/NIGMS NIH HHS/United States
- P30 CA062203/CA/NCI NIH HHS/United States
- DP2 AT010376/AT/NCCIH NIH HHS/United States
- GT11549/HHMI/Howard Hughes Medical Institute/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

