Model-based analysis uncovers mutations altering autophagy selectivity in human cancer
- PMID: 34059679
- PMCID: PMC8166871
- DOI: 10.1038/s41467-021-23539-5
Model-based analysis uncovers mutations altering autophagy selectivity in human cancer
Abstract
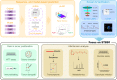
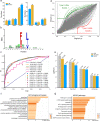
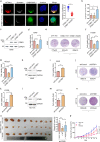
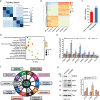
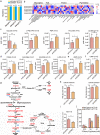
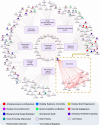
Autophagy can selectively target protein aggregates, pathogens, and dysfunctional organelles for the lysosomal degradation. Aberrant regulation of autophagy promotes tumorigenesis, while it is far less clear whether and how tumor-specific alterations result in autophagic aberrance. To form a link between aberrant autophagy selectivity and human cancer, we establish a computational pipeline and prioritize 222 potential LIR (LC3-interacting region) motif-associated mutations (LAMs) in 148 proteins. We validate LAMs in multiple proteins including ATG4B, STBD1, EHMT2 and BRAF that impair their interactions with LC3 and autophagy activities. Using a combination of transcriptomic, metabolomic and additional experimental assays, we show that STBD1, a poorly-characterized protein, inhibits tumor growth via modulating glycogen autophagy, while a patient-derived W203C mutation on LIR abolishes its cancer inhibitory function. This work suggests that altered autophagy selectivity is a frequently-used mechanism by cancer cells to survive during various stresses, and provides a framework to discover additional autophagy-related pathways that influence carcinogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
iCAL: a new pipeline to investigate autophagy selectivity and cancer.Autophagy. 2021 Jul;17(7):1799-1801. doi: 10.1080/15548627.2021.1939972. Epub 2021 Jun 28. Autophagy. 2021. PMID: 34176435 Free PMC article.
Similar articles
-
iCAL: a new pipeline to investigate autophagy selectivity and cancer.Autophagy. 2021 Jul;17(7):1799-1801. doi: 10.1080/15548627.2021.1939972. Epub 2021 Jun 28. Autophagy. 2021. PMID: 34176435 Free PMC article.
-
Starch-binding domain-containing protein 1 (Stbd1) and glycogen metabolism: Identification of the Atg8 family interacting motif (AIM) in Stbd1 required for interaction with GABARAPL1.Biochem Biophys Res Commun. 2011 Sep 30;413(3):420-5. doi: 10.1016/j.bbrc.2011.08.106. Epub 2011 Aug 27. Biochem Biophys Res Commun. 2011. PMID: 21893048 Free PMC article.
-
NIMA-related kinase 9-mediated phosphorylation of the microtubule-associated LC3B protein at Thr-50 suppresses selective autophagy of p62/sequestosome 1.J Biol Chem. 2020 Jan 31;295(5):1240-1260. doi: 10.1074/jbc.RA119.010068. Epub 2019 Dec 19. J Biol Chem. 2020. PMID: 31857374 Free PMC article.
-
Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors.J Mol Biol. 2020 Jan 3;432(1):80-103. doi: 10.1016/j.jmb.2019.07.016. Epub 2019 Jul 13. J Mol Biol. 2020. PMID: 31310766 Review.
-
Impaired autophagy and APP processing in Alzheimer's disease: The potential role of Beclin 1 interactome.Prog Neurobiol. 2013 Jul-Aug;106-107:33-54. doi: 10.1016/j.pneurobio.2013.06.002. Epub 2013 Jul 1. Prog Neurobiol. 2013. PMID: 23827971 Review.
Cited by
-
The mechanisms and roles of selective autophagy in mammals.Nat Rev Mol Cell Biol. 2023 Mar;24(3):167-185. doi: 10.1038/s41580-022-00542-2. Epub 2022 Oct 27. Nat Rev Mol Cell Biol. 2023. PMID: 36302887 Review.
-
ROS generation attenuates the anti-cancer effect of CPX on cervical cancer cells by inducing autophagy and inhibiting glycophagy.Redox Biol. 2022 Jul;53:102339. doi: 10.1016/j.redox.2022.102339. Epub 2022 May 17. Redox Biol. 2022. PMID: 35636017 Free PMC article.
-
Cryo-EM structure of the Mon1-Ccz1-RMC1 complex reveals molecular basis of metazoan RAB7A activation.Proc Natl Acad Sci U S A. 2023 May 30;120(22):e2301725120. doi: 10.1073/pnas.2301725120. Epub 2023 May 22. Proc Natl Acad Sci U S A. 2023. PMID: 37216550 Free PMC article.
-
AlphaFold2-multimer guided high-accuracy prediction of typical and atypical ATG8-binding motifs.PLoS Biol. 2023 Feb 8;21(2):e3001962. doi: 10.1371/journal.pbio.3001962. eCollection 2023 Feb. PLoS Biol. 2023. PMID: 36753519 Free PMC article.
-
iCAL: a new pipeline to investigate autophagy selectivity and cancer.Autophagy. 2021 Jul;17(7):1799-1801. doi: 10.1080/15548627.2021.1939972. Epub 2021 Jun 28. Autophagy. 2021. PMID: 34176435 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

