Genome-wide identification of the BASS gene family in four Gossypium species and functional characterization of GhBASSs against salt stress
- PMID: 34059742
- PMCID: PMC8166867
- DOI: 10.1038/s41598-021-90740-3
Genome-wide identification of the BASS gene family in four Gossypium species and functional characterization of GhBASSs against salt stress
Abstract
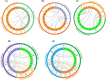
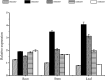
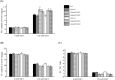
Bile acid sodium symporter (BASS) family proteins encode a class of sodium/solute symporters. Even though the sodium transporting property of BASSs in mammals was well studied, their sodium transportability and functional roles in plant salt tolerance remained largely unknown. Here, BASS family members from 4 cotton species, as well as 30 other species were identified. Then, they were designated as members of BASS1 to BASS5 subfamilies according to their sequence similarity and phylogenetic relationships. There were 8, 11, 16 and 18 putative BASS genes in four cotton species. While whole-genome duplications (WGD) and segmental duplications rendered the expansion of the BASS gene family in cotton, BASS gene losses occurred in the tetraploid cotton during the evolution from diploids to allotetraploids. Concerning functional characterizations, the transcript profiling of GhBASSs revealed that they not only preferred tissue-specific expression but also were differently induced by various stressors and phytohormones. Gene silencing and overexpression experiments showed that GhBASS1 and GhBASS3 positively regulated, whereas GhBASS2, GhBASS4 and GhBASS5 negatively regulated plant salt tolerance. Taken together, BASS family genes have evolved before the divergence from the common ancestor of prokaryotes and eukaryotes, and GhBASSs are plastidial sodium-dependent metabolite co-transporters that can influence plant salt tolerance.
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Schleiff U. Analysis of water supply of plants under saline soil conditions and conclusions for research on crop salt tolerance. J. Agron. Crop Sci. 2008;194:1–8. doi: 10.1111/j.1439-037X.2007.00290.x. - DOI
-
- Gorji T, Tanik A, Sertel E. Soil salinity prediction, monitoring and mapping using modern technologies. Procedia Earth Planet. Sci. 2015;15:507–512. doi: 10.1016/j.proeps.2015.08.062. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

