The effects of combined environmental factors on the intestinal flora of mice based on ground simulation experiments
- PMID: 34059794
- PMCID: PMC8166921
- DOI: 10.1038/s41598-021-91077-7
The effects of combined environmental factors on the intestinal flora of mice based on ground simulation experiments
Abstract
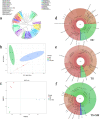
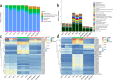
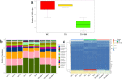
The composition and function of intestinal microbial communities are important for human health. However, these intestinal floras are sensitive to changes in the environment. Adverse changes to intestinal flora can affect the health of astronauts, resulting in difficulties in implementing space missions. We randomly divided mice into three groups and placed each group in either a normal environment, simulated microgravity environment or a combined effects environment, which included simulated microgravity, low pressure and noise. Fecal samples of the mice were collected for follow-up analysis based on metagenomics technology. With the influence of different space environmental factors, the species composition at the phylum and genus levels were significantly affected by the combined effects environment, especially the abundance of the Firmicutes and Bacteroidetes. Furthermore, screening was conducted to identify biomarkers that could be regarded as environmental markers. And there have also been some noticeable changes in the function of intestinal floras. Moreover, the abundance of antibiotic resistance genes (ARGs) was also found to be changed under different environmental conditions, such as bacitracin and vancomycin. The combined effects environment could significantly affect the species composition, function, and the expression of ARGs of intestinal flora of mice which may provide a theoretical basis for space medical supervision and healthcare.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Intestinal microbiota contributes to colonic epithelial changes in simulated microgravity mouse model.FASEB J. 2017 Aug;31(8):3695-3709. doi: 10.1096/fj.201700034R. Epub 2017 May 11. FASEB J. 2017. PMID: 28495755
-
Space Environmental Factor Impacts upon Murine Colon Microbiota and Mucosal Homeostasis.PLoS One. 2015 Jun 17;10(6):e0125792. doi: 10.1371/journal.pone.0125792. eCollection 2015. PLoS One. 2015. PMID: 26083373 Free PMC article.
-
Simulated space environmental factors of weightlessness, noise and low atmospheric pressure differentially affect the diurnal rhythm and the gut microbiome.Life Sci Space Res (Amst). 2024 Feb;40:115-125. doi: 10.1016/j.lssr.2023.09.006. Epub 2023 Sep 22. Life Sci Space Res (Amst). 2024. PMID: 38245336
-
Cardiovascular changes under the microgravity environment and the gut microbiome.Life Sci Space Res (Amst). 2024 Feb;40:89-96. doi: 10.1016/j.lssr.2023.09.003. Epub 2023 Sep 9. Life Sci Space Res (Amst). 2024. PMID: 38245353 Review.
-
The effects of microgravity on the digestive system and the new insights it brings to the life sciences.Life Sci Space Res (Amst). 2020 Nov;27:74-82. doi: 10.1016/j.lssr.2020.07.009. Epub 2020 Jul 30. Life Sci Space Res (Amst). 2020. PMID: 34756233 Review.
Cited by
-
Molecular Mechanism of Microgravity-Induced Intestinal Flora Dysbiosis on the Abnormalities of Liver and Brain Metabolism.Int J Mol Sci. 2025 Mar 27;26(7):3094. doi: 10.3390/ijms26073094. Int J Mol Sci. 2025. PMID: 40243802 Free PMC article.
-
Effects of Bifidobacterium lactis BLa80 on fecal and mucosal flora and stem cell factor/c-kit signaling pathway in simulated microgravity rats.World J Gastroenterol. 2025 Jan 7;31(1):96199. doi: 10.3748/wjg.v31.i1.96199. World J Gastroenterol. 2025. PMID: 39777246 Free PMC article.
-
Long-term simulated microgravity alters gut microbiota and metabolome in mice.Front Microbiol. 2023 Mar 24;14:1100747. doi: 10.3389/fmicb.2023.1100747. eCollection 2023. Front Microbiol. 2023. PMID: 37032862 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

