A single-center pilot study in Malaysia on the clinical utility of whole-exome sequencing for inborn errors of immunity
- PMID: 34060650
- PMCID: PMC8506128
- DOI: 10.1111/cei.13626
A single-center pilot study in Malaysia on the clinical utility of whole-exome sequencing for inborn errors of immunity
Abstract
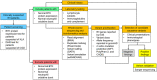
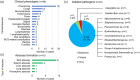
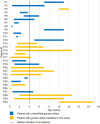
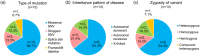
Primary immunodeficiency diseases refer to inborn errors of immunity (IEI) that affect the normal development and function of the immune system. The phenotypical and genetic heterogeneity of IEI have made their diagnosis challenging. Hence, whole-exome sequencing (WES) was employed in this pilot study to identify the genetic etiology of 30 pediatric patients clinically diagnosed with IEI. The potential causative variants identified by WES were validated using Sanger sequencing. Genetic diagnosis was attained in 46.7% (14 of 30) of the patients and categorized into autoinflammatory disorders (n = 3), diseases of immune dysregulation (n = 3), defects in intrinsic and innate immunity (n = 3), predominantly antibody deficiencies (n = 2), combined immunodeficiencies with associated and syndromic features (n = 2) and immunodeficiencies affecting cellular and humoral immunity (n = 1). Of the 15 genetic variants identified, two were novel variants. Genetic findings differed from the provisional clinical diagnoses in seven cases (50.0%). This study showed that WES enhances the capacity to diagnose IEI, allowing more patients to receive appropriate therapy and disease management.
Keywords: bioinformatics analysis; genetic diagnosis; genetic variant; inborn errors of immunity; whole-exome sequencing.
© 2021 The Authors. Clinical & Experimental Immunology published by John Wiley & Sons Ltd on behalf of British Society for Immunology.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures
Similar articles
-
Genetic Evaluation of the Patients with Clinically Diagnosed Inborn Errors of Immunity by Whole Exome Sequencing: Results from a Specialized Research Center for Immunodeficiency in Türkiye.J Clin Immunol. 2024 Jul 2;44(7):157. doi: 10.1007/s10875-024-01759-w. J Clin Immunol. 2024. PMID: 38954121 Free PMC article.
-
Phenotypic heterogeneity and genotypic spectrum of inborn errors of immunity identified through whole exome sequencing in a Thai patient cohort.Pediatr Allergy Immunol. 2022 Jan;33(1):e13701. doi: 10.1111/pai.13701. Epub 2021 Nov 28. Pediatr Allergy Immunol. 2022. PMID: 34796988
-
Clinical Utility of Whole Exome Sequencing and Targeted Panels for the Identification of Inborn Errors of Immunity in a Resource-Constrained Setting.Front Immunol. 2021 May 21;12:665621. doi: 10.3389/fimmu.2021.665621. eCollection 2021. Front Immunol. 2021. PMID: 34093558 Free PMC article.
-
Future of Therapy for Inborn Errors of Immunity.Clin Rev Allergy Immunol. 2022 Aug;63(1):75-89. doi: 10.1007/s12016-021-08916-8. Epub 2022 Jan 12. Clin Rev Allergy Immunol. 2022. PMID: 35020169 Free PMC article. Review.
-
Approach to genetic diagnosis of inborn errors of immunity through next-generation sequencing.Mol Immunol. 2021 Sep;137:57-66. doi: 10.1016/j.molimm.2021.06.018. Epub 2021 Jun 30. Mol Immunol. 2021. PMID: 34216999 Review.
Cited by
-
Non-infectious mixed cryoglobulinemia as a new clinical presentation of mutation in the gene encoding coatomer subunit alpha: a case report of two adult sisters.Front Immunol. 2024 Nov 15;15:1450048. doi: 10.3389/fimmu.2024.1450048. eCollection 2024. Front Immunol. 2024. PMID: 39620212 Free PMC article.
-
Diagnostic yield of next-generation sequencing in suspect primary immunodeficiencies diseases: a systematic review and meta-analysis.Clin Exp Med. 2024 Jun 18;24(1):131. doi: 10.1007/s10238-024-01392-2. Clin Exp Med. 2024. PMID: 38890201 Free PMC article.
-
Visual inspection reveals a novel pathogenic mutation in PKD1 missed by the variant caller in whole‑exome sequencing.Mol Med Rep. 2022 Dec;26(6):365. doi: 10.3892/mmr.2022.12882. Epub 2022 Oct 25. Mol Med Rep. 2022. PMID: 36281931 Free PMC article.
-
Revealing Chronic Granulomatous Disease in a Patient With Williams-Beuren Syndrome Using Whole Exome Sequencing.Front Immunol. 2021 Nov 4;12:778133. doi: 10.3389/fimmu.2021.778133. eCollection 2021. Front Immunol. 2021. PMID: 34804071 Free PMC article.
-
Human inborn errors of immunity underlying Talaromyces marneffei infections: a multicenter, retrospective cohort study.Front Immunol. 2025 Jan 22;16:1492000. doi: 10.3389/fimmu.2025.1492000. eCollection 2025. Front Immunol. 2025. PMID: 39911395 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

