Control of the Serine Integrase Reaction: Roles of the Coiled-Coil and Helix E Regions in DNA Site Synapsis and Recombination
- PMID: 34060907
- PMCID: PMC8297529
- DOI: 10.1128/JB.00703-20
Control of the Serine Integrase Reaction: Roles of the Coiled-Coil and Helix E Regions in DNA Site Synapsis and Recombination
Abstract
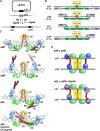
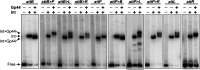
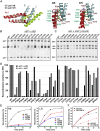
Bacteriophage serine integrases catalyze highly specific recombination reactions between defined DNA segments called att sites. These reactions are reversible depending upon the presence of a second phage-encoded directionality factor. The bipartite C-terminal DNA-binding region of integrases includes a recombinase domain (RD) connected to a zinc-binding domain (ZD), which contains a long flexible coiled-coil (CC) motif that extends away from the bound DNA. We directly show that the identities of the phage A118 integrase att sites are specified by the DNA spacing between the RD and ZD DNA recognition determinants, which in turn directs the relative trajectories of the CC motifs on each subunit of the att-bound integrase dimer. Recombination between compatible dimer-bound att sites requires minimal-length CC motifs and 14 residues surrounding the tip where the pairing of CC motifs between synapsing dimers occurs. Our alanine-scanning data suggest that molecular interactions between CC motif tips may differ in integrative (attP × attB) and excisive (attL × attR) recombination reactions. We identify mutations in 5 residues within the integrase oligomerization helix that control the remodeling of dimers into tetramers during synaptic complex formation. Whereas most of these gain-of-function mutants still require the CC motifs for synapsis, one mutant efficiently, but indiscriminately, forms synaptic complexes without the CC motifs. However, the CC motifs are still required for recombination, suggesting a function for the CC motifs after the initial assembly of the integrase synaptic tetramer. IMPORTANCE The robust and exquisitely regulated site-specific recombination reactions promoted by serine integrases are integral to the life cycle of temperate bacteriophage and, in the case of the A118 prophage, are an important virulence factor of Listeria monocytogenes. The properties of these recombinases have led to their repurposing into tools for genetic engineering and synthetic biology. In this report, we identify determinants regulating synaptic complex formation between correct DNA sites, including the DNA architecture responsible for specifying the identity of recombination sites, features of the unique coiled-coil structure on the integrase that are required to initiate synapsis, and amino acid residues on the integrase oligomerization helix that control the remodeling of synapsing dimers into a tetramer active for DNA strand exchange.
Keywords: Listeria monocytogenes; phage A118; serine recombinase; site-specific DNA recombination; synaptic complex.
Figures




Similar articles
-
Control of Recombination Directionality by the Listeria Phage A118 Protein Gp44 and the Coiled-Coil Motif of Its Serine Integrase.J Bacteriol. 2017 May 9;199(11):e00019-17. doi: 10.1128/JB.00019-17. Print 2017 Jun 1. J Bacteriol. 2017. PMID: 28289084 Free PMC article.
-
Coiled-coil interactions mediate serine integrase directionality.Nucleic Acids Res. 2017 Jul 7;45(12):7339-7353. doi: 10.1093/nar/gkx474. Nucleic Acids Res. 2017. PMID: 28549184 Free PMC article.
-
DNA cleavage is independent of synapsis during Streptomyces phage phiBT1 integrase-mediated site-specific recombination.J Mol Cell Biol. 2010 Oct;2(5):264-75. doi: 10.1093/jmcb/mjq025. J Mol Cell Biol. 2010. PMID: 20871112
-
Phage-encoded Serine Integrases and Other Large Serine Recombinases.Microbiol Spectr. 2015 Aug;3(4). doi: 10.1128/microbiolspec.MDNA3-0059-2014. Microbiol Spectr. 2015. PMID: 26350324 Review.
-
Site-specific recombination by phiC31 integrase and other large serine recombinases.Biochem Soc Trans. 2010 Apr;38(2):388-94. doi: 10.1042/BST0380388. Biochem Soc Trans. 2010. PMID: 20298189 Review.
Cited by
-
There and turn back again: the application of phage serine integrases in eukaryotic systems.Front Bioeng Biotechnol. 2025 Feb 24;13:1478413. doi: 10.3389/fbioe.2025.1478413. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40066361 Free PMC article. Review.
-
Large serine integrases utilise scavenged phage proteins as directionality cofactors.Nucleic Acids Res. 2025 Jan 24;53(3):gkaf050. doi: 10.1093/nar/gkaf050. Nucleic Acids Res. 2025. PMID: 39907112 Free PMC article.
-
Structural basis of directionality control in large serine integrases.bioRxiv [Preprint]. 2025 Jan 13:2025.01.03.631226. doi: 10.1101/2025.01.03.631226. bioRxiv. 2025. PMID: 39803483 Free PMC article. Preprint.
-
Protocol for the establishment of a serine integrase-based platform for functional validation of genetic switch controllers in eukaryotic cells.PLoS One. 2024 May 23;19(5):e0303999. doi: 10.1371/journal.pone.0303999. eCollection 2024. PLoS One. 2024. PMID: 38781126 Free PMC article.
-
A phage weaponizes a satellite recombinase to subvert viral restriction.Nucleic Acids Res. 2022 Oct 28;50(19):11138-11153. doi: 10.1093/nar/gkac845. Nucleic Acids Res. 2022. PMID: 36259649 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

