Evolution of VIM-1-Producing Klebsiella pneumoniae Isolates from a Hospital Outbreak Reveals the Genetic Bases of the Loss of the Urease-Positive Identification Character
- PMID: 34060914
- PMCID: PMC8269217
- DOI: 10.1128/mSystems.00244-21
Evolution of VIM-1-Producing Klebsiella pneumoniae Isolates from a Hospital Outbreak Reveals the Genetic Bases of the Loss of the Urease-Positive Identification Character
Abstract
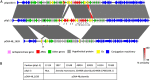
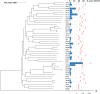
Outbreaks of carbapenemase-producing Klebsiella pneumoniae (CPKp) represent a major threat for hospitals. We molecularly characterized the first outbreak of VIM-1-producing K. pneumoniae in Spain, which raised fears about the spread of this strain or of the plasmid carrying blaVIM-1. Through in-depth genomic analysis of 18 isolates recovered between October 2005 and September 2007, we show that 17 ST39 isolates were clonal, whereas the last isolate had acquired the VIM-1 plasmid from the epidemic clone. The index isolate carried 31 antibiotic resistance genes (ARGs) and was resistant to almost all antibiotics tested. Later isolates further gained mutations in efflux pump regulators ramR and opxR, deletion of mgrB (colistin resistance), and frameshift mutations in ompK36 (β-lactam resistance) likely selected by antibiotic usage. Comparison with publicly available genome sequences and literature review revealed no sign of dissemination of this CPKp strain. However, the VIM-1 plasmid was found in diverse Enterobacterales species, although restricted to Spain. One isolate became urease negative following IS5075 transposition into ureC. Analysis of 9,755 K. pneumoniae genomes showed the same ureC::IS5075 insertion in 14.1% of the isolates and explained why urease activity is a variable identification trait for K. pneumoniae. Transposition into ureC results from the similarity of its 3' end and the terminal inverted repeats of Tn21-like transposons, the targets of IS5075 and related insertion sequences (ISs). As these transposons frequently carry ARGs, this might explain the frequent chromosomal invasion by these ISs and ureC inactivation in multidrug-resistant isolates. IMPORTANCE Evolution of multidrug-resistant bacterial pathogens occurs at multiple scales, in the patient, locally in the hospital, or more globally. Some mutations or gene acquisitions, for instance in response to antibiotic treatment, may be restricted to a single patient due to their high fitness cost. However, some events are more general. By analyzing the evolution of a hospital-acquired multidrug-resistant K. pneumoniae strain producing the carbapenemase VIM-1, we showed a likely environmental source in the hospital and identified mutations contributing to a further decrease in antibiotic susceptibility. By combining the genomic analysis of this outbreak with literature data and genome sequences available in databases, we showed that the VIM-1 plasmid has been acquired by different Enterobacterales but is endemic only in Spain. We also discovered that urease loss in K. pneumoniae results from the specific transposition of an IS element into the ureC gene and was more frequent in fluoroquinolone-resistant isolates and those carrying a carbapenemase gene.
Keywords: Klebsiella pneumoniae; carbapenemase; drug resistance evolution; insertion sequence; mobile genetic elements; urease.
Figures




Similar articles
-
Early OXA-48-Producing Enterobacterales Isolates Recovered in a Spanish Hospital Reveal a Complex Introduction Dominated by Sequence Type 11 (ST11) and ST405 Klebsiella pneumoniae Clones.mSphere. 2020 Apr 8;5(2):e00080-20. doi: 10.1128/mSphere.00080-20. mSphere. 2020. PMID: 32269151 Free PMC article.
-
A ten-year surveillance study of carbapenemase-producing Klebsiella pneumoniae in a tertiary care Greek university hospital: predominance of KPC- over VIM- or NDM-producing isolates.J Med Microbiol. 2016 Mar;65(3):240-246. doi: 10.1099/jmm.0.000217. Epub 2015 Dec 23. J Med Microbiol. 2016. PMID: 26698320
-
Microbiological and molecular characteristics of carbapenemase-producing Klebsiella pneumoniae endemic in a tertiary Greek hospital during 2004-2010.Euro Surveill. 2012 Feb 16;17(7):20088. Euro Surveill. 2012. PMID: 22370015
-
Evaluation of the Therapeutic Outcomes of Antibiotic Regimen Against Carbapenemase-Producing Klebsiella pneumoniae: A Systematic Review and Meta-Analysis.Front Pharmacol. 2021 Nov 4;12:597907. doi: 10.3389/fphar.2021.597907. eCollection 2021. Front Pharmacol. 2021. PMID: 34803661 Free PMC article. Review.
-
Plasmid Dissemination in Multispecies Carbapenemase-Producing Enterobacterales Outbreaks Involving Clinical and Environmental Strains: A Narrative Review.Microorganisms. 2025 Apr 2;13(4):810. doi: 10.3390/microorganisms13040810. Microorganisms. 2025. PMID: 40284646 Free PMC article. Review.
Cited by
-
Longitudinal analysis within one hospital in sub-Saharan Africa over 20 years reveals repeated replacements of dominant clones of Klebsiella pneumoniae and stresses the importance to include temporal patterns for vaccine design considerations.Genome Med. 2024 May 6;16(1):67. doi: 10.1186/s13073-024-01342-3. Genome Med. 2024. PMID: 38711148 Free PMC article.
-
Multidrug-resistant hypervirulent Klebsiella pneumoniae: an evolving superbug.Future Microbiol. 2025 Apr;20(6):499-511. doi: 10.1080/17460913.2025.2482478. Epub 2025 Mar 26. Future Microbiol. 2025. PMID: 40135944 Review.
-
Genetic Characterization of Carbapenem-Resistant Klebsiella pneumoniae Clinical Isolates in a Tertiary Hospital in Greece, 2018-2022.Antibiotics (Basel). 2023 May 28;12(6):976. doi: 10.3390/antibiotics12060976. Antibiotics (Basel). 2023. PMID: 37370295 Free PMC article.
-
Virulence factors in carbapenem-resistant hypervirulent Klebsiella pneumoniae.Front Microbiol. 2023 Nov 30;14:1325077. doi: 10.3389/fmicb.2023.1325077. eCollection 2023. Front Microbiol. 2023. PMID: 38098668 Free PMC article. Review.
-
Genomic analysis of the international high-risk clonal lineage Klebsiella pneumoniae sequence type 395.Genome Med. 2023 Feb 13;15(1):9. doi: 10.1186/s13073-023-01159-6. Genome Med. 2023. PMID: 36782220 Free PMC article.
References
-
- David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H. 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi:10.1038/s41564-019-0492-8. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
