Host Species and Geography Differentiate Honeybee Gut Bacterial Communities by Changing the Relative Contribution of Community Assembly Processes
- PMID: 34061602
- PMCID: PMC8262996
- DOI: 10.1128/mBio.00751-21
Host Species and Geography Differentiate Honeybee Gut Bacterial Communities by Changing the Relative Contribution of Community Assembly Processes
Abstract
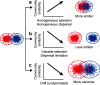
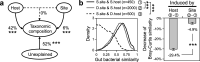
Honeybee gut microbiota modulates the health and fitness of honeybees, the ecologically and economically important pollinators and honey producers. However, which processes drive the assembly and shift of honeybee gut microbiota remains unknown. To explore the patterns of honeybee gut bacterial communities across host species and geographical sites and the relative contribution of different processes (i.e., homogeneous selection, variable selection, homogeneous dispersal, dispersal limitation, and an undominated process) in driving the patterns, two honeybee species (Apis cerana and Apis mellifera) were sampled from five geographically distant sites along a latitudinal gradient, followed by gut bacterial 16S rRNA gene sequencing. The gut bacterial communities differed significantly between A. cerana and A. mellifera, which was driven by the interhost dispersal limitation associated with the long-term coevolution between hosts and their prokaryotic symbionts. A. mellifera harbored more diverse but less varied gut bacterial communities than A. cerana due to the dominant role of homogeneous selection in converging A. mellifera intestinal communities. For each honeybee species, the gut bacterial communities differed across geographical sites, with individuals from lower latitudes harboring higher diversity; also, there was significant decay of gut community similarity against geographic distance. The geographical variation of honeybee gut bacterial communities was mainly driven by an undominated process (e.g., stochastic drift) rather than variable selection or dispersal limitation. This study elucidates that variations in host and geography alter the relative contribution of different processes in assembling honeybee gut microbiota and, thus, provides insights into the mechanisms underlying honeybee gut microbial shifts across evolutionary time. IMPORTANCE Honeybees provide crucial pollination services and valuable apiarian products. The symbiotic intestinal communities facilitate honeybee health and fitness by promoting nutrient assimilation, detoxifying toxins, and resisting pathogens. Thus, understanding the processes that govern honeybee gut bacterial communities is imperative for better managing gut microbiota to improve honeybee health. However, little is known about the processes driving the assembly and shift of honeybee gut bacterial communities. This study quantitatively deciphers the relative importance of selection, dispersal, and undominated processes in governing the assembly of honeybee gut bacterial communities and explores how their relative importance varies across biological and spatial scales. Our study provides new insights into the mechanisms underlying the maintenance and shift of honeybee gut microbiota.
Keywords: bacterial community; ecological processes; geography; gut microbiota; honeybee; host; neutrality; selection; stochasticity.
Figures




References
-
- Anderson KE, Sheehan TH, Eckholm BJ, Mott BM, DeGrandi-Hoffman G. 2011. An emerging paradigm of colony health: microbial balance of the honey bee and hive (Apis mellifera). Insect Soc 58:431–444. doi: 10.1007/s00040-011-0194-6. - DOI
-
- Dai PL, Zhou W, Zhang J, Jiang W-Y, Wang Q, Cui H-J, Sun J-H, Wu Y-Y, Zhou T. 2012. The effects of Bt Cry1Ah toxin on worker honeybees (Apis mellifera ligustica and Apis cerana cerana). Apidologie 43:384–391. doi: 10.1007/s13592-011-0103-z. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

