Do monogenic inborn errors of immunity cause susceptibility to severe COVID-19?
- PMID: 34061775
- PMCID: PMC8279660
- DOI: 10.1172/JCI149459
Do monogenic inborn errors of immunity cause susceptibility to severe COVID-19?
Abstract
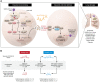
The SARS-CoV-2 virus, which causes COVID-19, has been associated globally with substantial morbidity and mortality. Numerous reports over the past year have described the clinical and immunological profiles of COVID-19 patients, and while some trends have emerged for risk stratification, they do not provide a complete picture. Therefore, efforts are ongoing to identify genetic susceptibility factors of severe disease. In this issue of the JCI, Povysil et al. performed a large, multiple-country study, sequencing genomes from patients with mild and severe COVID-19, along with population controls. Contrary to previous reports, the authors observed no enrichment of predicted loss-of-function variants in genes in the type I interferon pathway, which might predispose to severe disease. These studies suggest that more evidence is needed to substantiate the hypothesis for a genetic immune predisposition to severe COVID-19, and highlights the importance of considering experimental design when implicating a monogenic basis for severe disease.
Conflict of interest statement
Figures
Comment on
-
Inborn errors of type I IFN immunity in patients with life-threatening COVID-19.Science. 2020 Oct 23;370(6515):eabd4570. doi: 10.1126/science.abd4570. Epub 2020 Sep 24. Science. 2020. PMID: 32972995 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

