TRIP13 modulates protein deubiquitination and accelerates tumor development and progression of B cell malignancies
- PMID: 34061780
- PMCID: PMC8279583
- DOI: 10.1172/JCI146893
TRIP13 modulates protein deubiquitination and accelerates tumor development and progression of B cell malignancies
Abstract
Multiple myeloma (MM), a terminally differentiated B cell malignancy, remains difficult to cure. Understanding the molecular mechanisms underlying the progression of MM may identify therapeutic targets and lead to a fundamental shift in treatment of the disease. Deubiquitination, like ubiquitination, is a highly regulated process, implicated in almost every cellular process. Multiple deubiquitinating enzymes (DUBs) have been identified, but their regulation is poorly defined. Here, we determined that TRIP13 increases cellular deubiquitination. Overexpression of TRIP13 in mice and cultured cells resulted in excess cellular deubiquitination by enhancing the association of the DUB USP7 with its substrates. We show that TRIP13 is an oncogenic protein because it accelerates B cell tumor development in transgenic mice. TRIP13-induced resistance to proteasome inhibition can be overcome by a USP7 inhibitor in vitro and in vivo. These findings suggest that TRIP13 expression plays a critical role in B cell lymphoma and MM by regulating deubiquitination of critical oncogenic (NEK2) and tumor suppressor (PTEN, p53) proteins. High TRIP13 identifies a high-risk patient group amenable to adjuvant anti-USP7 therapy.
Keywords: Drug therapy; Hematology; Oncogenes; Ubiquitin-proteosome system.
Conflict of interest statement
Figures





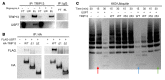
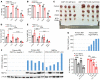

References
-
- Hanamura I, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108(5):1724–1732. doi: 10.1182/blood-2006-03-009910. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

