Implications of diffusion and time-varying morphogen gradients for the dynamic positioning and precision of bistable gene expression boundaries
- PMID: 34061823
- PMCID: PMC8195430
- DOI: 10.1371/journal.pcbi.1008589
Implications of diffusion and time-varying morphogen gradients for the dynamic positioning and precision of bistable gene expression boundaries
Abstract
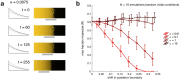
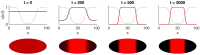
The earliest models for how morphogen gradients guide embryonic patterning failed to account for experimental observations of temporal refinement in gene expression domains. Following theoretical and experimental work in this area, dynamic positional information has emerged as a conceptual framework to discuss how cells process spatiotemporal inputs into downstream patterns. Here, we show that diffusion determines the mathematical means by which bistable gene expression boundaries shift over time, and therefore how cells interpret positional information conferred from morphogen concentration. First, we introduce a metric for assessing reproducibility in boundary placement or precision in systems where gene products do not diffuse, but where morphogen concentrations are permitted to change in time. We show that the dynamics of the gradient affect the sensitivity of the final pattern to variation in initial conditions, with slower gradients reducing the sensitivity. Second, we allow gene products to diffuse and consider gene expression boundaries as propagating wavefronts with velocity modulated by local morphogen concentration. We harness this perspective to approximate a PDE model as an ODE that captures the position of the boundary in time, and demonstrate the approach with a preexisting model for Hunchback patterning in fruit fly embryos. We then propose a design that employs antiparallel morphogen gradients to achieve accurate boundary placement that is robust to scaling. Throughout our work we draw attention to tradeoffs among initial conditions, boundary positioning, and the relative timescales of network and gradient evolution. We conclude by suggesting that mathematical theory should serve to clarify not just our quantitative, but also our intuitive understanding of patterning processes.
Conflict of interest statement
The author has declared that no competing interests exist.
Figures




References
-
- Briscoe J. Understanding Pattern Formation in Embryos: Experiment, Theory, and Simulation. Journal of Computational Biology. 2019;26(0):1–7. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

