Rare variants regulate expression of nearby individual genes in multiple tissues
- PMID: 34061836
- PMCID: PMC8195400
- DOI: 10.1371/journal.pgen.1009596
Rare variants regulate expression of nearby individual genes in multiple tissues
Abstract
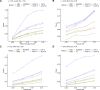
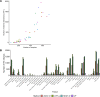
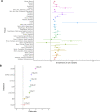
The rapid decrease in sequencing cost has enabled genetic studies to discover rare variants associated with complex diseases and traits. Once this association is identified, the next step is to understand the genetic mechanism of rare variants on how the variants influence diseases. Similar to the hypothesis of common variants, rare variants may affect diseases by regulating gene expression, and recently, several studies have identified the effects of rare variants on gene expression using heritability and expression outlier analyses. However, identifying individual genes whose expression is regulated by rare variants has been challenging due to the relatively small sample size of expression quantitative trait loci studies and statistical approaches not optimized to detect the effects of rare variants. In this study, we analyze whole-genome sequencing and RNA-seq data of 681 European individuals collected for the Genotype-Tissue Expression (GTEx) project (v8) to identify individual genes in 49 human tissues whose expression is regulated by rare variants. To improve statistical power, we develop an approach based on a likelihood ratio test that combines effects of multiple rare variants in a nonlinear manner and has higher power than previous approaches. Using GTEx data, we identify many genes regulated by rare variants, and some of them are only regulated by rare variants and not by common variants. We also find that genes regulated by rare variants are enriched for expression outliers and disease-causing genes. These results suggest the regulatory effects of rare variants, which would be important in interpreting associations of rare variants with complex traits.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Tissue specific regulation of transcription in endometrium and association with disease.Hum Reprod. 2020 Feb 29;35(2):377-393. doi: 10.1093/humrep/dez279. Hum Reprod. 2020. PMID: 32103259 Free PMC article.
-
Leveraging allelic imbalance to refine fine-mapping for eQTL studies.PLoS Genet. 2019 Dec 13;15(12):e1008481. doi: 10.1371/journal.pgen.1008481. eCollection 2019 Dec. PLoS Genet. 2019. PMID: 31834882 Free PMC article.
-
Quantifying the contribution of sequence variants with regulatory and evolutionary significance to 34 bovine complex traits.Proc Natl Acad Sci U S A. 2019 Sep 24;116(39):19398-19408. doi: 10.1073/pnas.1904159116. Epub 2019 Sep 9. Proc Natl Acad Sci U S A. 2019. PMID: 31501319 Free PMC article.
-
Identifying rare variants associated with complex traits via sequencing.Curr Protoc Hum Genet. 2013 Jul;Chapter 1:Unit 1.26. doi: 10.1002/0471142905.hg0126s78. Curr Protoc Hum Genet. 2013. PMID: 23853079 Free PMC article. Review.
-
The Role of Noncoding Variants in Heritable Disease.Trends Genet. 2020 Nov;36(11):880-891. doi: 10.1016/j.tig.2020.07.004. Epub 2020 Jul 31. Trends Genet. 2020. PMID: 32741549 Review.
Cited by
-
Deleterious Mutations and the Rare Allele Burden on Rice Gene Expression.Mol Biol Evol. 2022 Sep 1;39(9):msac193. doi: 10.1093/molbev/msac193. Mol Biol Evol. 2022. PMID: 36073358 Free PMC article.
-
Deep sequencing of proteotoxicity modifier genes uncovers a Presenilin-2/beta-amyloid-actin genetic risk module shared among alpha-synucleinopathies.bioRxiv [Preprint]. 2024 Mar 7:2024.03.03.583145. doi: 10.1101/2024.03.03.583145. bioRxiv. 2024. PMID: 38496508 Free PMC article. Preprint.
-
Single-cell genomics meets human genetics.Nat Rev Genet. 2023 Aug;24(8):535-549. doi: 10.1038/s41576-023-00599-5. Epub 2023 Apr 21. Nat Rev Genet. 2023. PMID: 37085594 Free PMC article. Review.
-
Common and rare genetic variants show network convergence for a majority of human traits.medRxiv [Preprint]. 2025 Jun 28:2025.06.27.25330419. doi: 10.1101/2025.06.27.25330419. medRxiv. 2025. PMID: 40666368 Free PMC article. Preprint.
-
Transcriptome analysis provides critical answers to the "variants of uncertain significance" conundrum.Hum Mutat. 2022 Nov;43(11):1590-1608. doi: 10.1002/humu.24394. Epub 2022 May 18. Hum Mutat. 2022. PMID: 35510381 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

